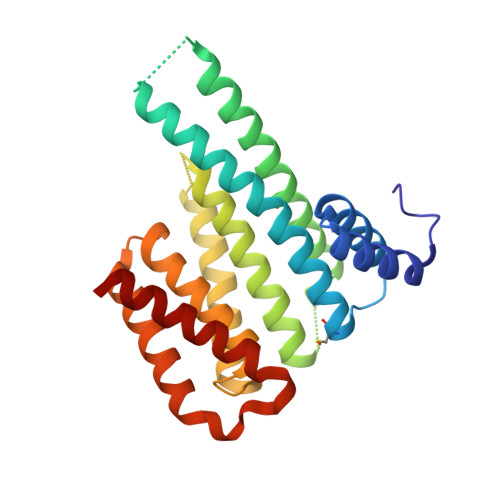
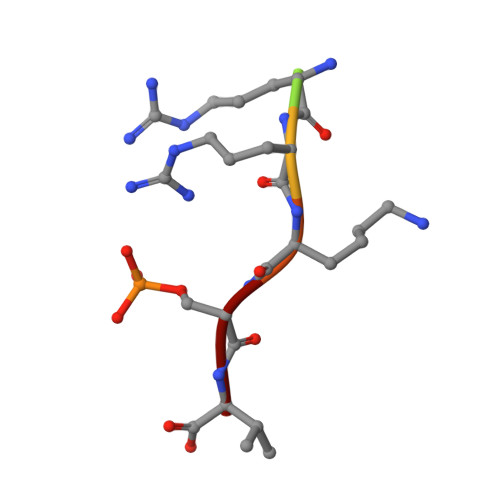
Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Andrei, S.A., de Vink, P., Sijbesma, E., Han, L., Brunsveld, L., Kato, N., Ottmann, C., Higuchi, Y.(2018) Angew Chem Int Ed Engl 57: 13470-13474
- PubMed: 30025189
- DOI: https://doi.org/10.1002/anie.201806584
- Primary Citation of Related Structures:
6GHP - PubMed Abstract:
The natural product family of fusicoccanes are stabilizers of 14-3-3 mediated protein-protein interactions (PPIs), some of which possess antitumor activity. In this study, the first use of molecular dynamics (MD) to rationally design PPI stabilizers with increased potency is presented. Synthesis of a focused library, with subsequent characterization by fluorescence polarization, mutational studies, and X-ray crystallography confirmed the power of the MD-based design approach, revealing the potential for an additional hydrogen bond with the 14-3-3 protein to lead to significantly increased potency. Additionally, these compounds exert their action in a cellular environment with increased potency. The newly found polar interaction could provide an anchoring point for new small-molecule PPI stabilizers. These results facilitate the development of fusicoccanes towards drugs or tool compounds, as well as allowing the study of the fundamental principles behind PPI stabilization.
- Laboratory of Chemical Biology and Institute for Complex Molecular Systems, Department of Biomedical Engineering, Eindhoven University of Technology, Den Dolech 2, 5612, AZ, Eindhoven, The Netherlands.
Organizational Affiliation: