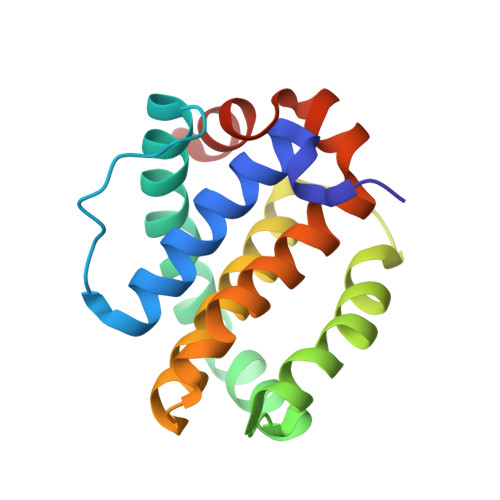
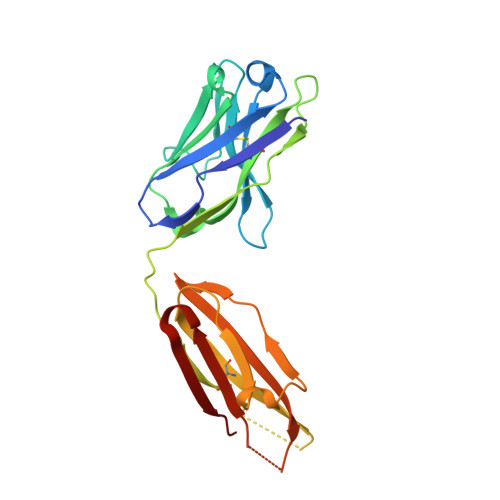
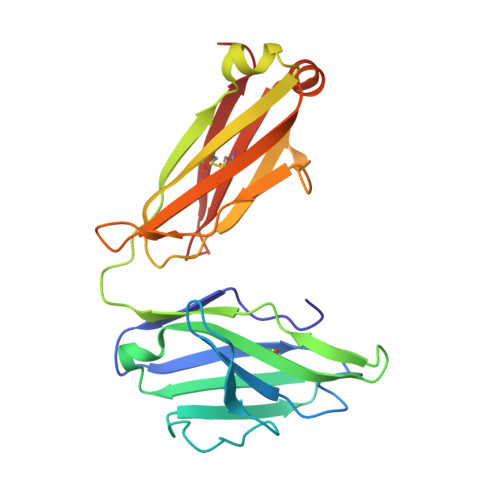
Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Tron, A.E., Belmonte, M.A., Adam, A., Aquila, B.M., Boise, L.H., Chiarparin, E., Cidado, J., Embrey, K.J., Gangl, E., Gibbons, F.D., Gregory, G.P., Hargreaves, D., Hendricks, J.A., Johannes, J.W., Johnstone, R.W., Kazmirski, S.L., Kettle, J.G., Lamb, M.L., Matulis, S.M., Nooka, A.K., Packer, M.J., Peng, B., Rawlins, P.B., Robbins, D.W., Schuller, A.G., Su, N., Yang, W., Ye, Q., Zheng, X., Secrist, J.P., Clark, E.A., Wilson, D.M., Fawell, S.E., Hird, A.W.(2018) Nat Commun 9: 5341-5341
- PubMed: 30559424
- DOI: https://doi.org/10.1038/s41467-018-07551-w
- Primary Citation of Related Structures:
6FS0, 6FS1, 6FS2 - PubMed Abstract:
Mcl-1 is a member of the Bcl-2 family of proteins that promotes cell survival by preventing induction of apoptosis in many cancers. High expression of Mcl-1 causes tumorigenesis and resistance to anticancer therapies highlighting the potential of Mcl-1 inhibitors as anticancer drugs. Here, we describe AZD5991, a rationally designed macrocyclic molecule with high selectivity and affinity for Mcl-1 currently in clinical development. Our studies demonstrate that AZD5991 binds directly to Mcl-1 and induces rapid apoptosis in cancer cells, most notably myeloma and acute myeloid leukemia, by activating the Bak-dependent mitochondrial apoptotic pathway. AZD5991 shows potent antitumor activity in vivo with complete tumor regression in several models of multiple myeloma and acute myeloid leukemia after a single tolerated dose as monotherapy or in combination with bortezomib or venetoclax. Based on these promising data, a Phase I clinical trial has been launched for evaluation of AZD5991 in patients with hematological malignancies (NCT03218683).
- Oncology, IMED Biotech Unit, AstraZeneca, Waltham, MA, 02451, USA.
Organizational Affiliation: