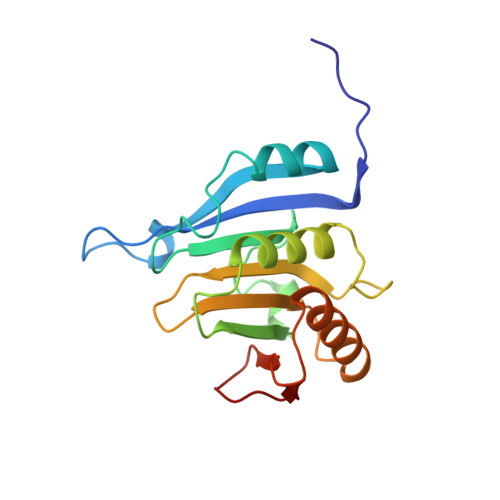
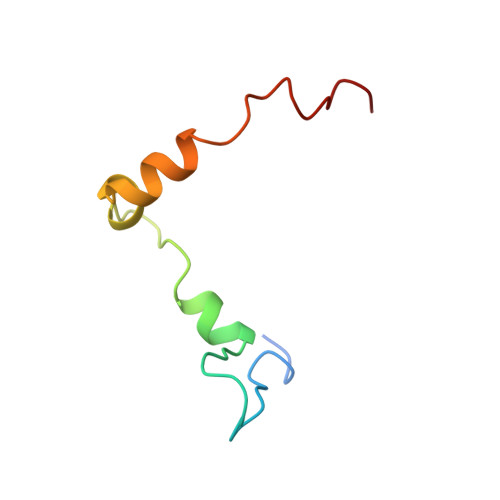
Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Gruner, S., Weber, R., Peter, D., Chung, M.Y., Igreja, C., Valkov, E., Izaurralde, E.(2018) Nucleic Acids Res 46: 6893-6908
- PubMed: 30053226
- DOI: https://doi.org/10.1093/nar/gky542
- Primary Citation of Related Structures:
6FBZ, 6FC0, 6FC1, 6FC2, 6FC3 - PubMed Abstract:
The interaction of the eukaryotic initiation factor 4G (eIF4G) with the cap-binding protein eIF4E initiates cap-dependent translation and is regulated by the 4E-binding proteins (4E-BPs), which compete with eIF4G to repress translation. Metazoan eIF4G and 4E-BPs interact with eIF4E via canonical and non-canonical motifs that bind to the dorsal and lateral surface of eIF4E in a bipartite recognition mode. However, previous studies pointed to mechanistic differences in how fungi and metazoans regulate protein synthesis. We present crystal structures of the yeast eIF4E bound to two yeast 4E-BPs, p20 and Eap1p, as well as crystal structures of a fungal eIF4E-eIF4G complex. We demonstrate that the core principles of molecular recognition of eIF4E are in fact highly conserved among translational activators and repressors in eukaryotes. Finally, we reveal that highly specialized structural motifs do exist and serve to modulate the affinity of protein-protein interactions that regulate cap-dependent translation initiation in fungi.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, D-72076 Tübingen, Germany.
Organizational Affiliation: