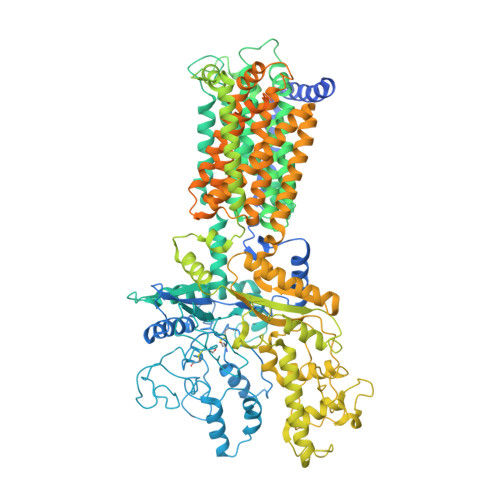
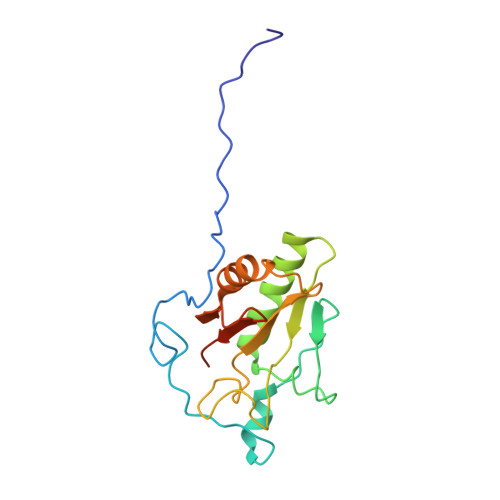
Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex.
Qi, X., Schmiege, P., Coutavas, E., Li, X.(2018) Science 362
- PubMed: 30139912
- DOI: https://doi.org/10.1126/science.aas8843
- Primary Citation of Related Structures:
6E1H - PubMed Abstract:
Aberrant Hedgehog (HH) signaling leads to various types of cancer and birth defects. N-terminally palmitoylated HH initiates signaling by binding its receptor Patched-1 (PTCH1). A recent 1:1 PTCH1-HH complex structure visualized a palmitate-mediated binding site on HH, which was inconsistent with previous studies that implied a distinct, calcium-mediated binding site for PTCH1 and HH co-receptors. Our 3.5-angstrom resolution cryo-electron microscopy structure of native Sonic Hedgehog (SHH-N) in complex with PTCH1 at a physiological calcium concentration reconciles these disparate findings and demonstrates that one SHH-N molecule engages both epitopes to bind two PTCH1 receptors in an asymmetric manner. Functional assays using PTCH1 or SHH-N mutants that disrupt the individual interfaces illustrate that simultaneous engagement of both interfaces is required for efficient signaling in cells.
- Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: