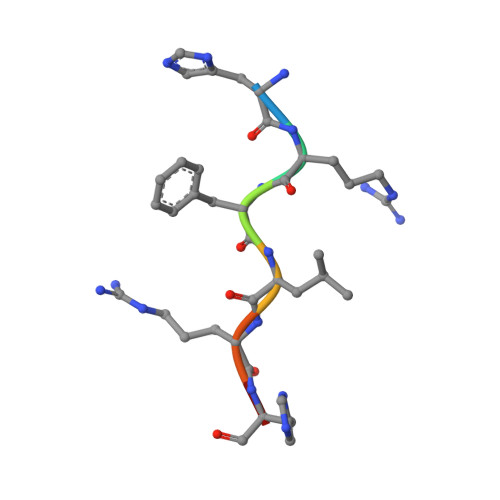
A previously undescribed hexapeptide His-Arg-Phe-Leu-Arg-His-NH2from amphibian skin secretion shows CO2and metal biding affinities.
Pires, D.A.T., Arake, L.M.R., Silva, L.P., Lopez-Castillo, A., Prates, M.V., Nascimento, C.J., Bloch, C.(2018) Peptides 106: 37-44
- PubMed: 29933027
- DOI: https://doi.org/10.1016/j.peptides.2018.06.003
- Primary Citation of Related Structures:
6CXN, 6CXP, 6CXQ, 6CXR - PubMed Abstract:
A previously undescribed six residues long peptide His-Arg-Phe-Leu-Arg-His was identified and purified from the skin secretion of the amphibian Phyllomedusa centralis. A synthetic analogue carboxyamidated HRFLRH-NH 2 showed structural changes induced by CO 2 and metal ions in aqueous solution when analyzed by NMR. The present work reports NMR structures for the carboxyamidated hexapeptide in the presence CO 2 , Zn 2+ and Cd 2+ , suggesting possible affinity regions on the polypeptide chain for each ligand. The NMR structures were optimized by DFT to identify probable biding sites of these species in the polypeptide structure. To our best knowledge, this is the first time that a putative CO 2 binding site is described on a peptide structure obtained in aqueous conditions, at room temperature.
- Instituto de Química, Universidade de Brasília (UnB), Brasília, Distrito Federal, Brazil; Departamento de Áreas Acadêmicas, Instituto Federal de Educação, Ciência e Tecnologia e Goiás (IFG), Luziânia, Goiás, Brazil.
Organizational Affiliation: