HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Morris, K.L., Buffalo, C.Z., Sturzel, C.M., Heusinger, E., Kirchhoff, F., Ren, X., Hurley, J.H.(2018) Cell 174: 659-671.e14
- PubMed: 30053425
- DOI: https://doi.org/10.1016/j.cell.2018.07.004
- Primary Citation of Related Structures:
6CM9, 6CRI, 6D83, 6D84, 6DFF - PubMed Abstract:
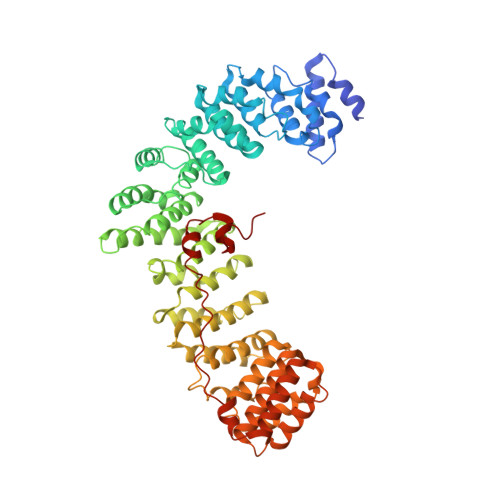
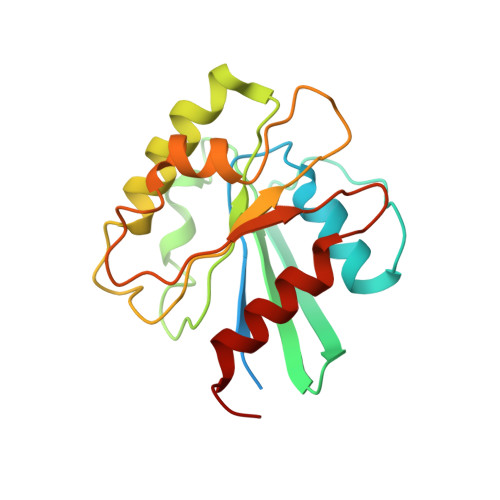
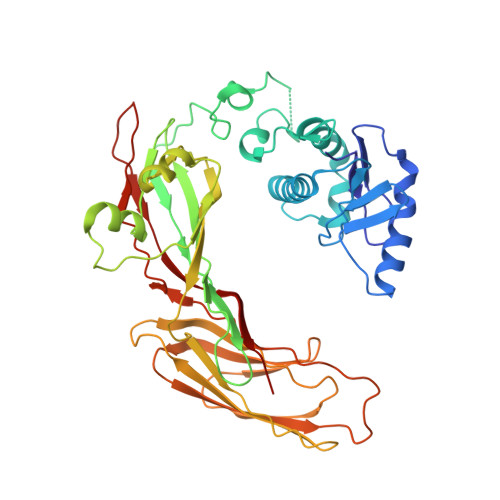
The HIV accessory protein Nef counteracts immune defenses by subverting coated vesicle pathways. The 3.7 Å cryo-EM structure of a closed trimer of the clathrin adaptor AP-1, the small GTPase Arf1, HIV-1 Nef, and the cytosolic tail of the restriction factor tetherin suggested a mechanism for inactivating tetherin by Golgi retention. The 4.3 Å structure of a mutant Nef-induced dimer of AP-1 showed how the closed trimer is regulated by the dileucine loop of Nef. HDX-MS and mutational analysis were used to show how cargo dynamics leads to alternative Arf1 trimerization, directing Nef targets to be either retained at the trans-Golgi or sorted to lysosomes. Phosphorylation of the NL4-3 M-Nef was shown to regulate AP-1 trimerization, explaining how O-Nefs lacking this phosphosite counteract tetherin but most M-Nefs do not. These observations show how the higher-order organization of a vesicular coat can be allosterically modulated to direct cargoes to distinct fates.
- Department of Molecular and Cell Biology and California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, CA 94720, USA.
Organizational Affiliation: