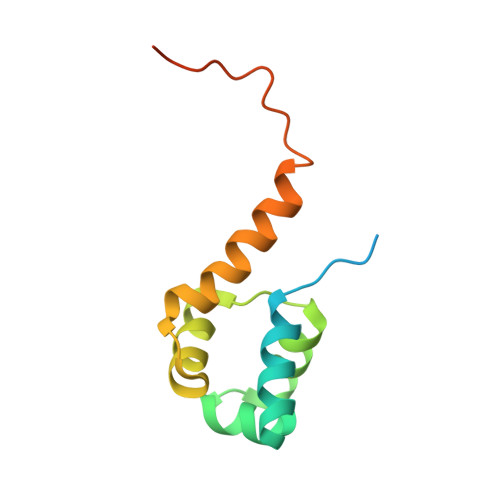
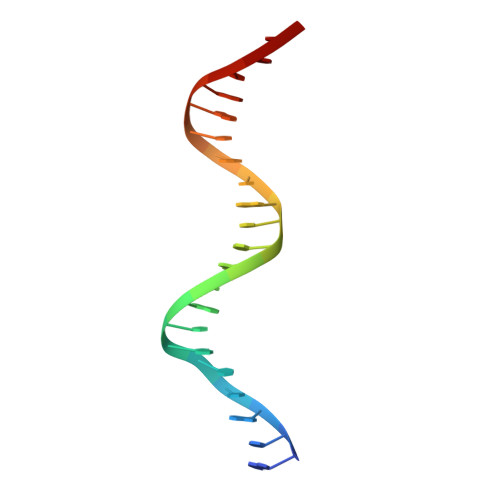
Structural basis of transcriptional regulation by the HigA antitoxin.
Schureck, M.A., Meisner, J., Hoffer, E.D., Wang, D., Onuoha, N., Ei Cho, S., Lollar 3rd, P., Dunham, C.M.(2019) Mol Microbiol 111: 1449-1462
- PubMed: 30793388
- DOI: https://doi.org/10.1111/mmi.14229
- Primary Citation of Related Structures:
6CF1, 6CHV - PubMed Abstract:
Bacterial toxin-antitoxin systems are important factors implicated in growth inhibition and plasmid maintenance. Type II toxin-antitoxin pairs are regulated at the transcriptional level by the antitoxin itself. Here, we examined how the HigA antitoxin regulates the expression of the Proteus vulgaris higBA toxin-antitoxin operon from the Rts1 plasmid. The HigBA complex adopts a unique architecture suggesting differences in its regulation as compared to classical type II toxin-antitoxin systems. We find that the C-terminus of the HigA antitoxin is required for dimerization and transcriptional repression. Further, the HigA structure reveals that the C terminus is ordered and does not transition between disorder-to-order states upon toxin binding. HigA residue Arg40 recognizes a TpG dinucleotide in higO2, an evolutionary conserved mode of recognition among prokaryotic and eukaryotic transcription factors. Comparison of the HigBA and HigA-higO2 structures reveals the distance between helix-turn-helix motifs of each HigA monomer increases by ~4 Å in order to bind to higO2. Consistent with these data, HigBA binding to each operator is twofold less tight than HigA alone. Together, these data show the HigB toxin does not act as a co-repressor suggesting potential novel regulation in this toxin-antitoxin system.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA, 30322, USA.
Organizational Affiliation: