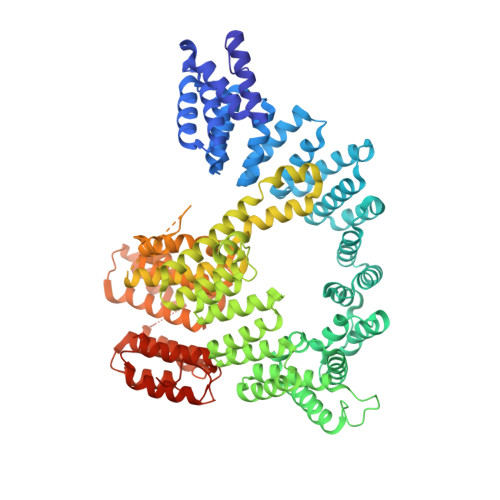
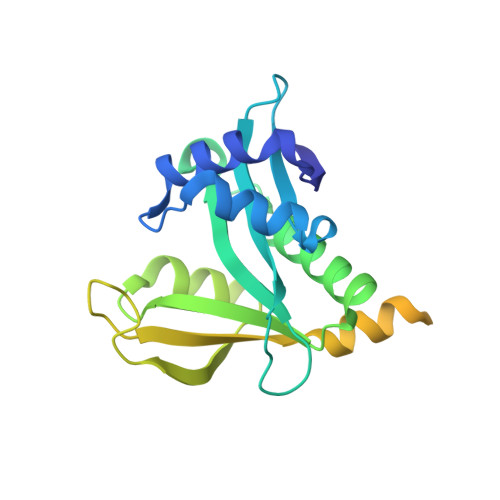
Structure of Human NatA and Its Regulation by the Huntingtin Interacting Protein HYPK.
Gottlieb, L., Marmorstein, R.(2018) Structure 26: 925
- PubMed: 29754825
- DOI: https://doi.org/10.1016/j.str.2018.04.003
- Primary Citation of Related Structures:
6C95, 6C9M - PubMed Abstract:
Co-translational N-terminal protein acetylation regulates many protein functions including degradation, folding, interprotein interactions, and targeting. Human NatA (hNatA), one of six conserved metazoan N-terminal acetyltransferases, contains Naa10 catalytic and Naa15 auxiliary subunits, and associates with the intrinsically disordered Huntingtin yeast two-hybrid protein K (HYPK). We report on the crystal structures of hNatA and hNatA/HYPK, and associated biochemical and enzymatic analyses. We demonstrate that hNatA contains unique features: a stabilizing inositol hexaphosphate (IP 6 ) molecule and a metazoan-specific Naa15 domain that mediates high-affinity HYPK binding. We find that HYPK harbors intrinsic hNatA-specific inhibitory activity through a bipartite structure: a ubiquitin-associated domain that binds a hNaa15 metazoan-specific region and an N-terminal loop-helix region that distorts the hNaa10 active site. We show that HYPK binding blocks hNaa50 targeting to hNatA, likely limiting Naa50 ribosome localization in vivo. These studies provide a model for metazoan NAT activity and HYPK regulation of N-terminal acetylation.
- Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA. Electronic address: lgott@sas.upenn.edu.
Organizational Affiliation: