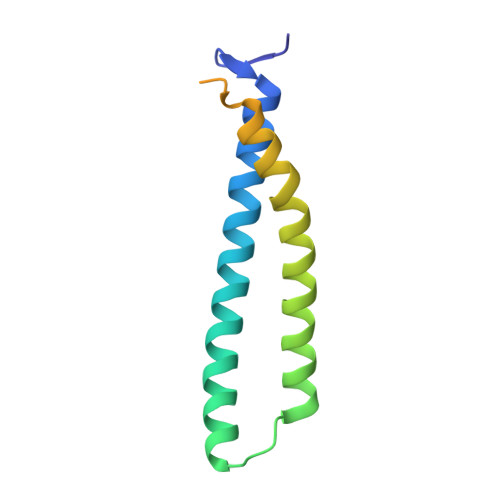
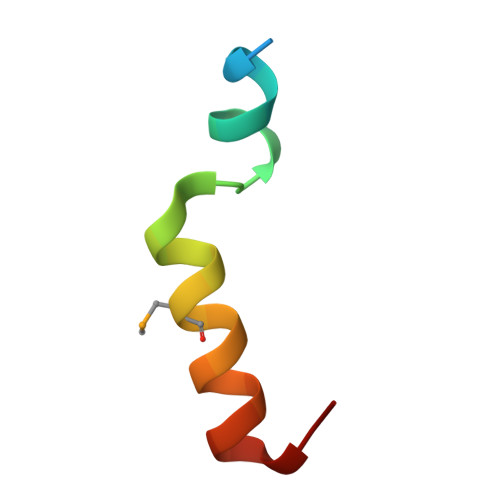
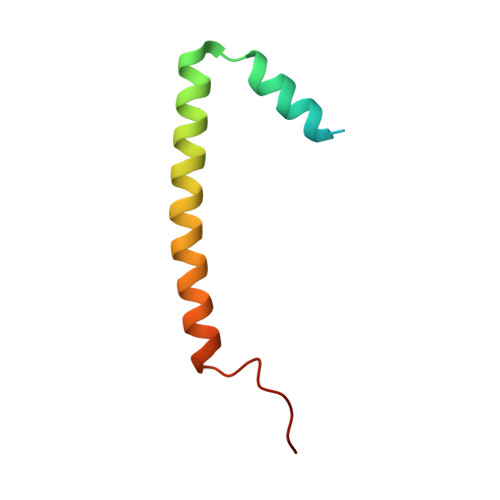
Structural mechanism of Myb-MuvB assembly.
Guiley, K.Z., Iness, A.N., Saini, S., Tripathi, S., Lipsick, J.S., Litovchick, L., Rubin, S.M.(2018) Proc Natl Acad Sci U S A 115: 10016-10021
- PubMed: 30224471
- DOI: https://doi.org/10.1073/pnas.1808136115
- Primary Citation of Related Structures:
6C48 - PubMed Abstract:
The MuvB transcriptional regulatory complex, which controls cell-cycle-dependent gene expression, cooperates with B-Myb to activate genes required for the G2 and M phases of the cell cycle. We have identified the domain in B-Myb that is essential for the assembly of the Myb-MuvB (MMB) complex. We determined a crystal structure that reveals how this B-Myb domain binds MuvB through the adaptor protein LIN52 and the scaffold protein LIN9. The structure and biochemical analysis provide an understanding of how oncogenic B-Myb is recruited to regulate genes required for cell-cycle progression, and the MMB interface presents a potential therapeutic target to inhibit cancer cell proliferation.
- Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA 95064.
Organizational Affiliation: