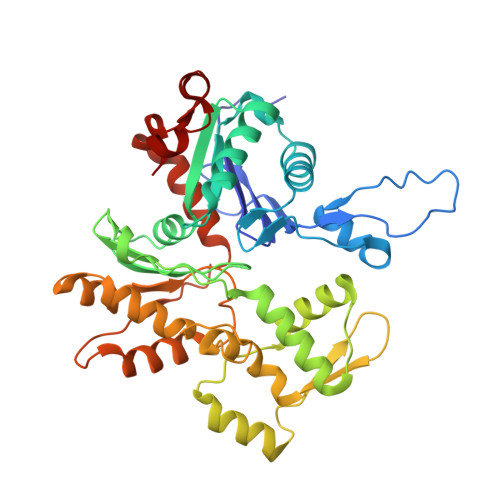
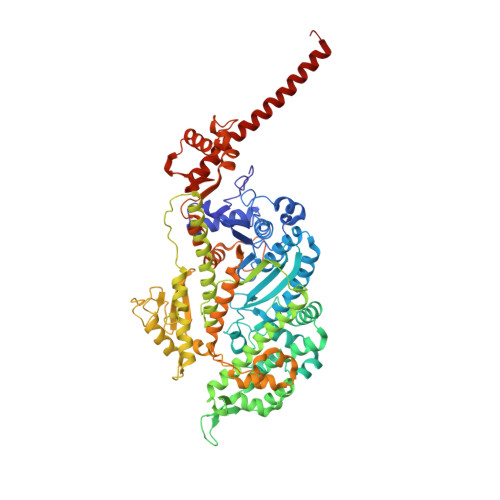
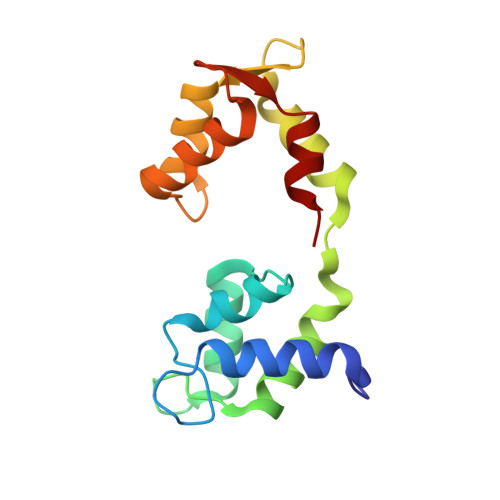
High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Mentes, A., Huehn, A., Liu, X., Zwolak, A., Dominguez, R., Shuman, H., Ostap, E.M., Sindelar, C.V.(2018) Proc Natl Acad Sci U S A 115: 1292-1297
- PubMed: 29358376
- DOI: https://doi.org/10.1073/pnas.1718316115
- Primary Citation of Related Structures:
5V7X, 6C1D, 6C1G, 6C1H - PubMed Abstract:
Myosins adjust their power outputs in response to mechanical loads in an isoform-dependent manner, resulting in their ability to dynamically adapt to a range of motile challenges. Here, we reveal the structural basis for force-sensing based on near-atomic resolution structures of one rigor and two ADP-bound states of myosin-IB (myo1b) bound to actin, determined by cryo-electron microscopy. The two ADP-bound states are separated by a 25° rotation of the lever. The lever of the first ADP state is rotated toward the pointed end of the actin filament and forms a previously unidentified interface with the N-terminal subdomain, which constitutes the upper half of the nucleotide-binding cleft. This pointed-end orientation of the lever blocks ADP release by preventing the N-terminal subdomain from the pivoting required to open the nucleotide binding site, thus revealing how myo1b is inhibited by mechanical loads that restrain lever rotation. The lever of the second ADP state adopts a rigor-like orientation, stabilized by class-specific elements of myo1b. We identify a role for this conformation as an intermediate in the ADP release pathway. Moreover, comparison of our structures with other myosins reveals structural diversity in the actomyosin binding site, and we reveal the high-resolution structure of actin-bound phalloidin, a potent stabilizer of filamentous actin. These results provide a framework to understand the spectrum of force-sensing capacities among the myosin superfamily.
- Pennsylvania Muscle Institute, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104.
Organizational Affiliation: