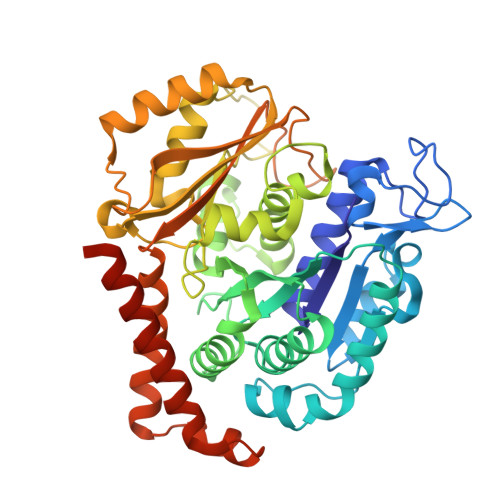
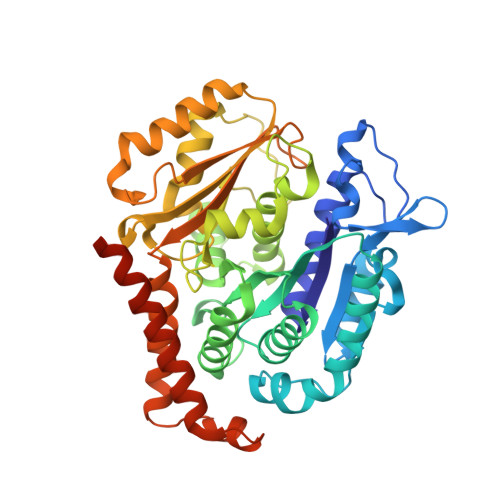
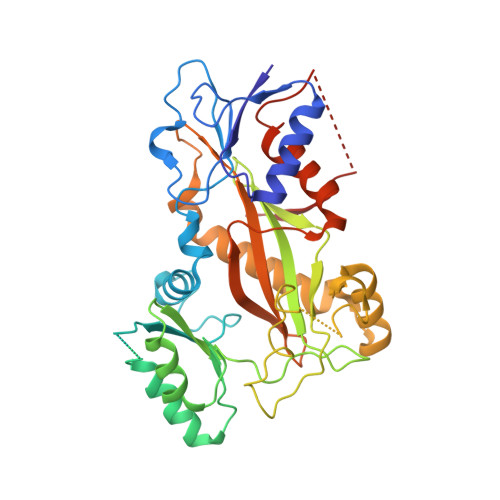
Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
Banerjee, S., Arnst, K.E., Wang, Y., Kumar, G., Deng, S., Yang, L., Li, G.B., Yang, J., White, S.W., Li, W., Miller, D.D.(2018) J Med Chem 61: 1704-1718
- PubMed: 29406710
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01858
- Primary Citation of Related Structures:
6BR1, 6BRF, 6BRY, 6BS2 - PubMed Abstract:
We report the design, synthesis, and biological evaluation of heterocyclic-fused pyrimidines as tubulin polymerization inhibitors targeting the colchicine binding site with significantly improved therapeutic index. Additionally, for the first time, we report high-resolution X-ray crystal structures for the best compounds in this scaffold, 4a, 4b, 6a, and 8b. These structures not only confirm their direct binding to the colchicine site in tubulin and reveal their detailed molecular interactions but also contrast the previously published proposed binding mode. Compounds 4a and 6a significantly inhibited tumor growth in an A375 melanoma xenograft model and were accompanied by elevated levels of apoptosis and disruption of tumor vasculature. Finally, we demonstrated that compound 4a significantly overcame clinically relevant multidrug resistance in a paclitaxel resistant PC-3/TxR prostate cancer xenograft model. Collectively, these studies provide preclinical and structural proof of concept to support the continued development of this scaffold as a new generation of tubulin inhibitors.
- Department of Pharmaceutical Sciences, College of Pharmacy, University of Tennessee Health Science Center , Memphis, Tennessee 38163, United States.
Organizational Affiliation: