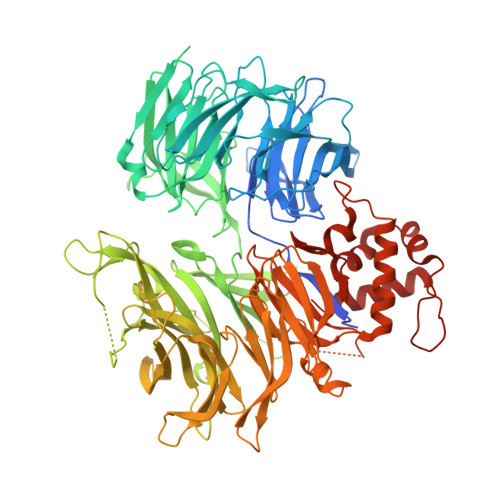
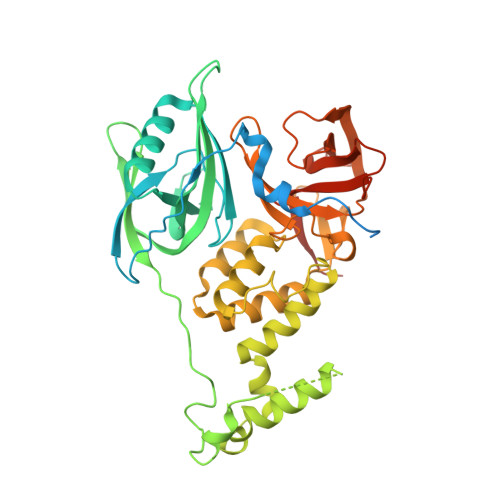
Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nowak, R.P., DeAngelo, S.L., Buckley, D., He, Z., Donovan, K.A., An, J., Safaee, N., Jedrychowski, M.P., Ponthier, C.M., Ishoey, M., Zhang, T., Mancias, J.D., Gray, N.S., Bradner, J.E., Fischer, E.S.(2018) Nat Chem Biol 14: 706-714
- PubMed: 29892083
- DOI: https://doi.org/10.1038/s41589-018-0055-y
- Primary Citation of Related Structures:
6BN7, 6BN8, 6BN9, 6BNB, 6BOY - PubMed Abstract:
Heterobifunctional small-molecule degraders that induce protein degradation through ligase-mediated ubiquitination have shown considerable promise as a new pharmacological modality. However, we currently lack a detailed understanding of the molecular basis for target recruitment and selectivity, which is critically required to enable rational design of degraders. Here we utilize a comprehensive characterization of the ligand-dependent CRBN-BRD4 interaction to demonstrate that binding between proteins that have not evolved to interact is plastic. Multiple X-ray crystal structures show that plasticity results in several distinct low-energy binding conformations that are selectively bound by ligands. We demonstrate that computational protein-protein docking can reveal the underlying interprotein contacts and inform the design of a BRD4 selective degrader that can discriminate between highly homologous BET bromodomains. Our findings that plastic interprotein contacts confer selectivity for ligand-induced protein dimerization provide a conceptual framework for the development of heterobifunctional ligands.
- Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA.
Organizational Affiliation: