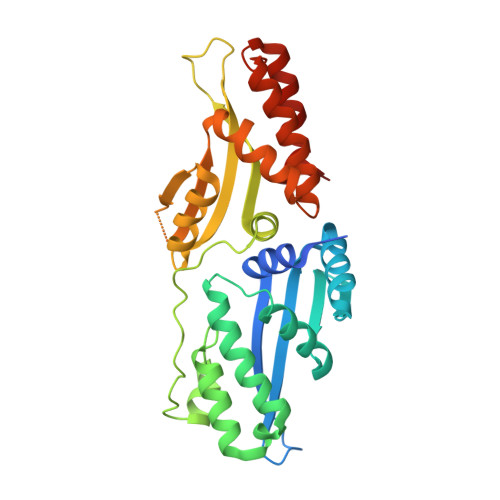
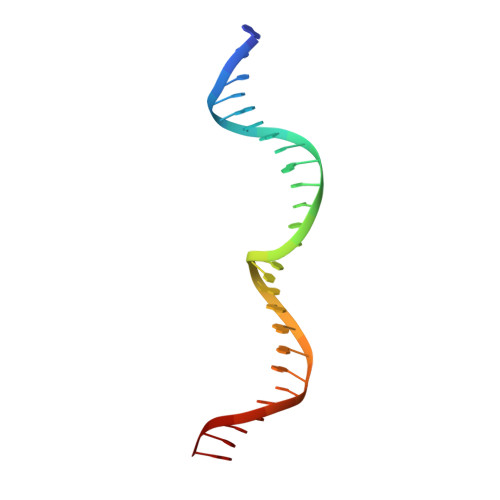
Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
McMurrough, T.A., Brown, C.M., Zhang, K., Hausner, G., Junop, M.S., Gloor, G.B., Edgell, D.R.(2018) Nucleic Acids Res 46: 11990-12007
- PubMed: 30357419
- DOI: https://doi.org/10.1093/nar/gky976
- Primary Citation of Related Structures:
6BCE, 6BCF, 6BCG, 6BCI, 6BCN, 6BCT - PubMed Abstract:
LAGLIDADG homing endonucleases (meganucleases) are site-specific mobile endonucleases that can be adapted for genome-editing applications. However, one problem when reprogramming meganucleases on non-native substrates is indirect readout of DNA shape and flexibility at the central 4 bases where cleavage occurs. To understand how the meganuclease active site regulates DNA cleavage, we used functional selections and deep sequencing to profile the fitness landscape of 1600 I-LtrI and I-OnuI active site variants individually challenged with 67 substrates with central 4 base substitutions. The wild-type active site was not optimal for cleavage on many substrates, including the native I-LtrI and I-OnuI targets. Novel combinations of active site residues not observed in known meganucleases supported activity on substrates poorly cleaved by the wild-type enzymes. Strikingly, combinations of E or D substitutions in the two metal-binding residues greatly influenced cleavage activity, and E184D variants had a broadened cleavage profile. Analyses of I-LtrI E184D and the wild-type proteins co-crystallized with the non-cognate AACC central 4 sequence revealed structural differences that correlated with kinetic constants for cleavage of individual DNA strands. Optimizing meganuclease active sites to enhance cleavage of non-native central 4 target sites is a straightforward addition to engineering workflows that will expand genome-editing applications.
- Department of Biochemistry, Schulich School of Medicine and Dentistry, Western University, London, ON, Canada, N6A 5C1.
Organizational Affiliation: