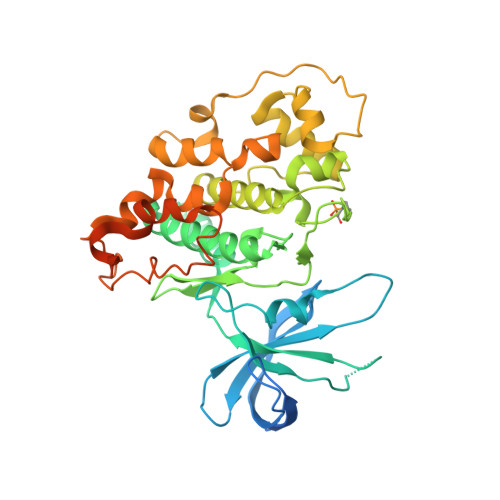
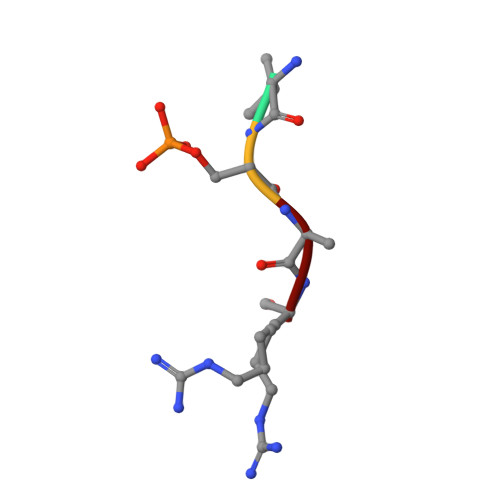
Synthesis, Binding Mode, and Antihyperglycemic Activity of Potent and Selective (5-Imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine Inhibitors of Glycogen Synthase Kinase 3.
Wagman, A.S., Boyce, R.S., Brown, S.P., Fang, E., Goff, D., Jansen, J.M., Le, V.P., Levine, B.H., Ng, S.C., Ni, Z.J., Nuss, J.M., Pfister, K.B., Ramurthy, S., Renhowe, P.A., Ring, D.B., Shu, W., Subramanian, S., Zhou, X.A., Shafer, C.M., Harrison, S.D., Johnson, K.W., Bussiere, D.E.(2017) J Med Chem 60: 8482-8514
- PubMed: 29016121
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00922
- Primary Citation of Related Structures:
6B8J - PubMed Abstract:
In an effort to identify new antidiabetic agents, we have discovered a novel family of (5-imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine analogues which are inhibitors of human glycogen synthase kinase 3 (GSK3). We developed efficient synthetic routes to explore a wide variety of substitution patterns and convergently access a diverse array of analogues. Compound 1 (CHIR-911, CT-99021, or CHIR-73911) emerged from an exploration of heterocycles at the C-5 position, phenyl groups at C-4, and a variety of differently substituted linker and aminopyridine moieties attached at the C-2 position. These compounds exhibited GSK3 IC 50 s in the low nanomolar range and excellent selectivity. They activate glycogen synthase in insulin receptor-expressing CHO-IR cells and primary rat hepatocytes. Evaluation of lead compounds 1 and 2 (CHIR-611 or CT-98014) in rodent models of type 2 diabetes revealed that single oral doses lowered hyperglycemia within 60 min, enhanced insulin-stimulated glucose transport, and improved glucose disposal without increasing insulin levels.
- Global Discovery Chemistry, Novartis Institutes for BioMedical Research , 5300 Chiron Way, Emeryville, California 94608, United States.
Organizational Affiliation: