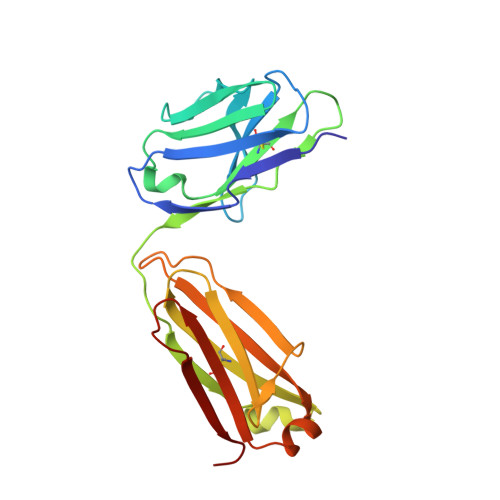
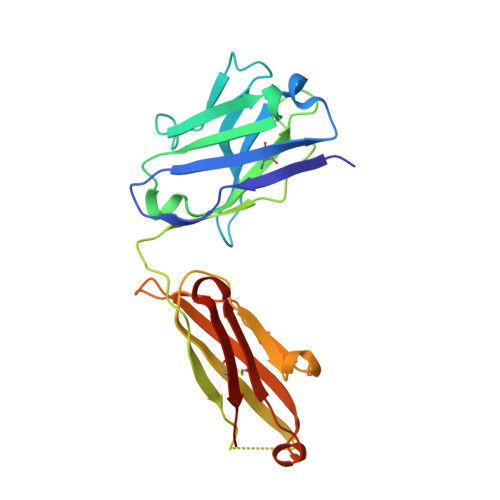
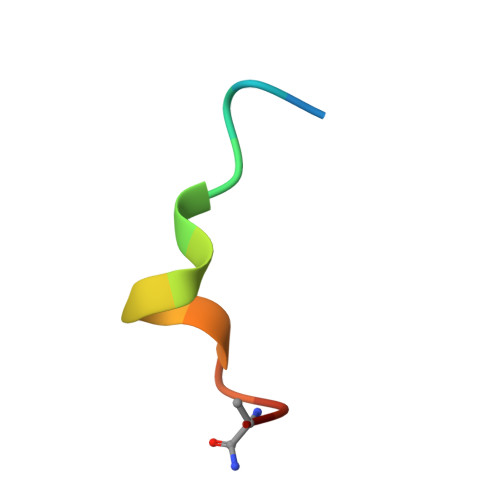
Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein.
Oyen, D., Torres, J.L., Wille-Reece, U., Ockenhouse, C.F., Emerling, D., Glanville, J., Volkmuth, W., Flores-Garcia, Y., Zavala, F., Ward, A.B., King, C.R., Wilson, I.A.(2017) Proc Natl Acad Sci U S A 114: E10438-E10445
- PubMed: 29138320
- DOI: https://doi.org/10.1073/pnas.1715812114
- Primary Citation of Related Structures:
6AXK, 6AXL - PubMed Abstract:
Acquired resistance against antimalarial drugs has further increased the need for an effective malaria vaccine. The current leading candidate, RTS,S, is a recombinant circumsporozoite protein (CSP)-based vaccine against Plasmodium falciparum that contains 19 NANP repeats followed by a thrombospondin repeat domain. Although RTS,S has undergone extensive clinical testing and has progressed through phase III clinical trials, continued efforts are underway to enhance its efficacy and duration of protection. Here, we determined that two monoclonal antibodies (mAbs 311 and 317), isolated from a recent controlled human malaria infection trial exploring a delayed fractional dose, inhibit parasite development in vivo by at least 97%. Crystal structures of antibody fragments (Fabs) 311 and 317 with an (NPNA) 3 peptide illustrate their different binding modes. Notwithstanding, one and three of the three NPNA repeats adopt similar well-defined type I β-turns with Fab311 and Fab317, respectively. Furthermore, to explore antibody binding in the context of P. falciparum CSP, we used negative-stain electron microscopy on a recombinant shortened CSP (rsCSP) construct saturated with Fabs. Both complexes display a compact rsCSP with multiple Fabs bound, with the rsCSP-Fab311 complex forming a highly organized helical structure. Together, these structural insights may aid in the design of a next-generation malaria vaccine.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037.
Organizational Affiliation: