A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1.
Vercoulen, Y., Kondo, Y., Iwig, J.S., Janssen, A., White, K.A., Amini, M., Barber, D.L., Kuriyan, J., Roose, J.P.(2017) Elife 6
- PubMed: 28952923
- DOI: https://doi.org/10.7554/eLife.29002
- Primary Citation of Related Structures:
6AXF, 6AXG - PubMed Abstract:
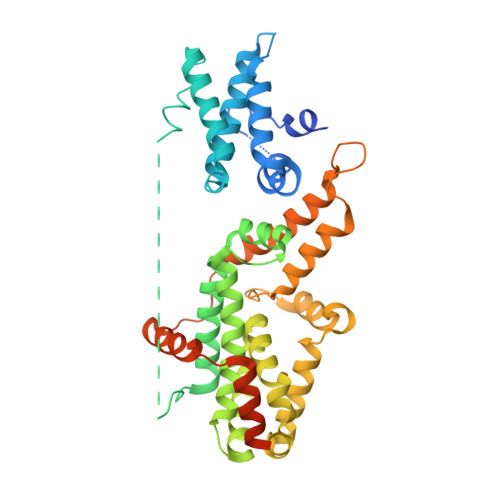
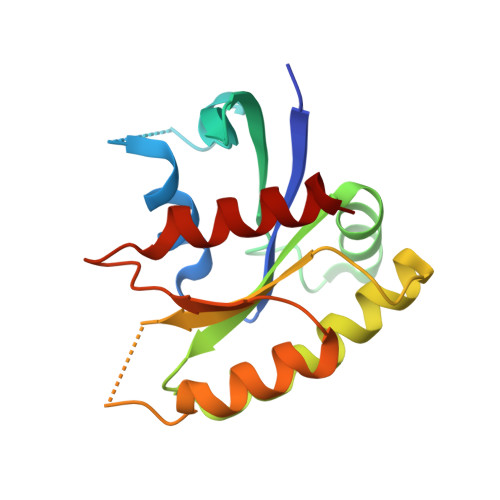
RasGRPs are guanine nucleotide exchange factors that are specific for Ras or Rap, and are important regulators of cellular signaling. Aberrant expression or mutation of RasGRPs results in disease. An analysis of RasGRP1 SNP variants led to the conclusion that the charge of His 212 in RasGRP1 alters signaling activity and plasma membrane recruitment, indicating that His 212 is a pH sensor that alters the balance between the inactive and active forms of RasGRP1. To understand the structural basis for this effect we compared the structure of autoinhibited RasGRP1, determined previously, to those of active RasGRP4:H-Ras and RasGRP2:Rap1b complexes. The transition from the autoinhibited to the active form of RasGRP1 involves the rearrangement of an inter-domain linker that displaces inhibitory inter-domain interactions. His 212 is located at the fulcrum of these conformational changes, and structural features in its vicinity are consistent with its function as a pH-dependent switch.
- Department of Anatomy, University of California, San Francisco, San Francisco, United States.
Organizational Affiliation: