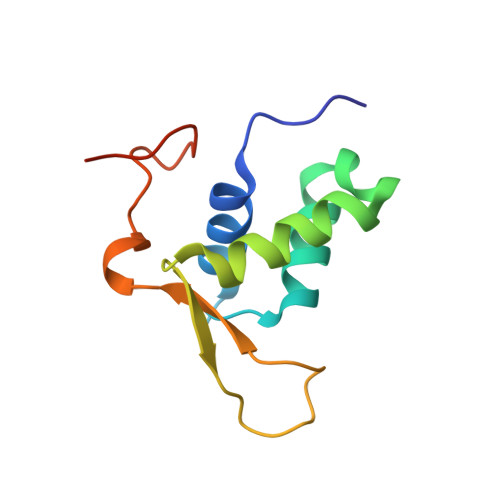
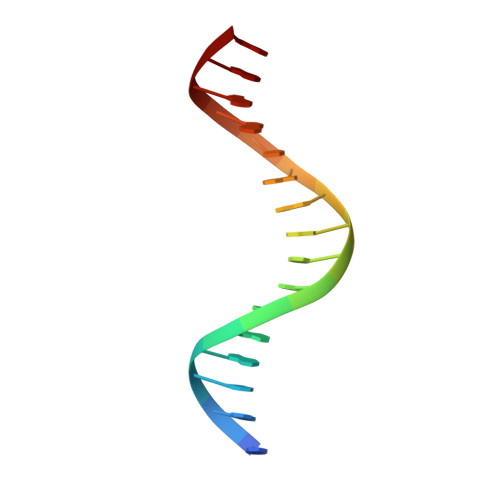
Structural basis for DNA recognition by FOXC2.
Chen, X., Wei, H., Li, J., Liang, X., Dai, S., Jiang, L., Guo, M., Qu, L., Chen, Z., Chen, L., Chen, Y.(2019) Nucleic Acids Res 47: 3752-3764
- PubMed: 30722065
- DOI: https://doi.org/10.1093/nar/gkz077
- Primary Citation of Related Structures:
6AKO, 6AKP - PubMed Abstract:
The FOXC family of transcription factors (FOXC1 and FOXC2) plays essential roles in the regulation of embryonic, ocular, and cardiac development. Mutations and abnormal expression of FOXC proteins are implicated in genetic diseases as well as cancer. In this study, we determined two crystal structures of the DNA-binding domain (DBD) of human FOXC2 protein, in complex with different DNA sites. The FOXC2-DBD adopts the winged-helix fold with helix H3 contributing to all the base specific contacts, while the N-terminus, wing 1, and the C-terminus of FOXC2-DBD all make additional contacts with the phosphate groups of DNA. Our structural, biochemical, and bioinformatics analyses allow us to revise the previously proposed DNA recognition mechanism and provide a model of DNA binding for the FOXC proteins. In addition, our structural analysis and accompanying biochemical assays provide a molecular basis for understanding disease-causing mutations in FOXC1 and FOXC2.
- NHC Key Laboratory of Cancer Proteomics and Laboratory of Structural Biology, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Organizational Affiliation: