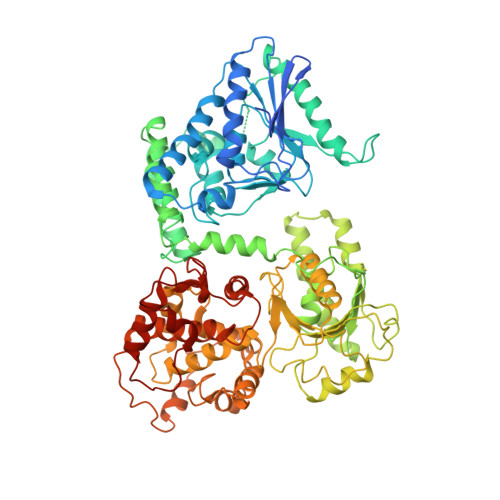
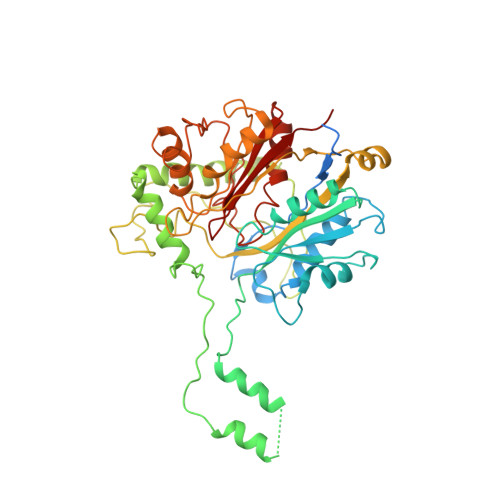
Cryo-EM structure of human mitochondrial trifunctional protein
Liang, K., Li, N., Wang, X., Dai, J., Liu, P., Wang, C., Chen, X.W., Gao, N., Xiao, J.(2018) Proc Natl Acad Sci U S A 115: 7039-7044
- PubMed: 29915090
- DOI: https://doi.org/10.1073/pnas.1801252115
- Primary Citation of Related Structures:
5ZQZ, 5ZRV - PubMed Abstract:
The mitochondrial trifunctional protein (TFP) catalyzes three reactions in the fatty acid β-oxidation process. Mutations in the two TFP subunits cause mitochondrial trifunctional protein deficiency and acute fatty liver of pregnancy that can lead to death. Here we report a 4.2-Å cryo-electron microscopy α2β2 tetrameric structure of the human TFP. The tetramer has a V-shaped architecture that displays a distinct assembly compared with the bacterial TFPs. A concave surface of the TFP tetramer interacts with the detergent molecules in the structure, suggesting that this region is involved in associating with the membrane. Deletion of a helical hairpin in TFPβ decreases its binding to the liposomes in vitro and reduces its membrane targeting in cells. Our results provide the structural basis for TFP function and have important implications for fatty acid oxidation related diseases.
- State Key Laboratory of Protein and Plant Gene Research, Peking University, 100871 Beijing, China.
Organizational Affiliation: