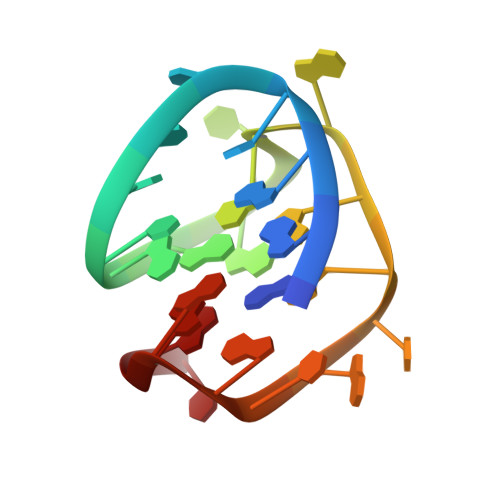
A putative G-quadruplex structure in the proximal promoter ofVEGFR-2has implications for drug design to inhibit tumor angiogenesis.
Liu, Y., Lan, W., Wang, C., Cao, C.(2018) J Biol Chem 293: 8947-8955
- PubMed: 29666187
- DOI: https://doi.org/10.1074/jbc.RA118.002666
- Primary Citation of Related Structures:
5ZEV - PubMed Abstract:
Tumor angiogenesis is mainly regulated by vascular endothelial growth factor (VEGF) produced by cancer cells. It is active on the endothelium via VEGF receptor 2 (VEGFR-2). G-quadruplexes are DNA secondary structures formed by guanine-rich sequences, for example, within gene promoters where they may contribute to transcriptional activity. The proximal promoter of VEGFR-2 contains a G-quadruplex, which has been suggested to interact with small molecules that inhibit VEGFR-2 expression and thereby tumor angiogenesis. However, its structure is not known. Here, we determined its NMR solution structure, which is composed of three stacked G-tetrads containing three syn guanines. The first guanine (G 1 ) is positioned within the central G-tetrad. We also observed that a noncanonical, V-shaped loop spans three G-tetrad planes, including no bridging nucleotides. A long and diagonal loop, which includes six nucleotides, connects reversal double chains. With a melting temperature of 54.51 °C, the scaffold of this quadruplex is stabilized by one G-tetrad plane stacking with one nonstandard bp, G 3 -C 8 , whose bases interact with each other through only one hydrogen bond. In summary, the NMR solution structure of the G-quadruplex in the proximal promoter region of the VEGFR-2 gene reported here has uncovered its key features as a potential anticancer drug target.
Organizational Affiliation:
From the State Key Laboratory of Bioorganic and Natural Product Chemistry, Center for Excellence in Molecular Synthesis, Shanghai Institute of Organic Chemistry and.