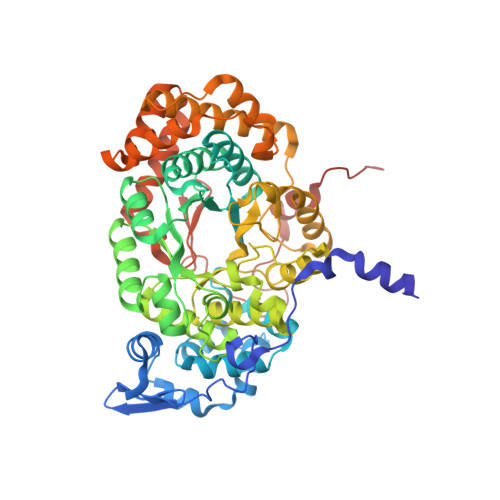
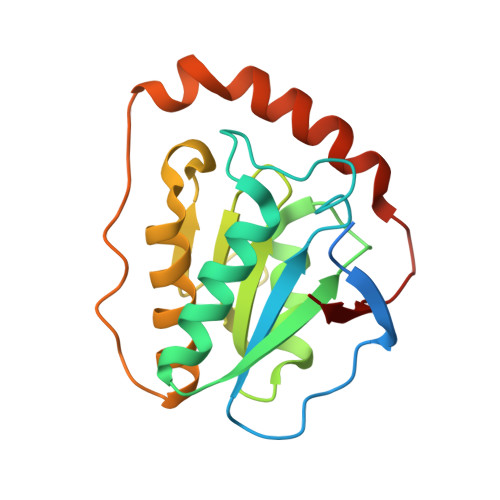
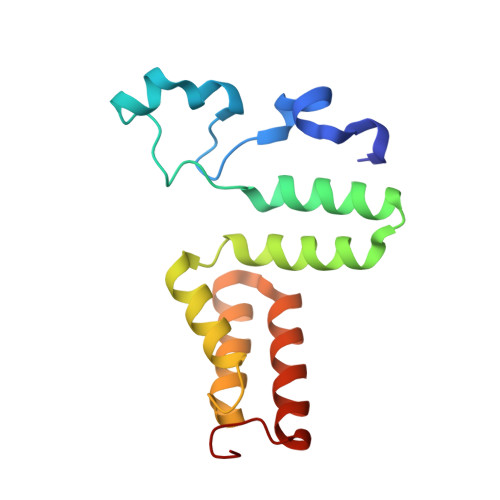
Direct Participation of a Peripheral Side Chain of a Corrin Ring in Coenzyme B12Catalysis.
Shibata, N., Sueyoshi, Y., Higuchi, Y., Toraya, T.(2018) Angew Chem Int Ed Engl 57: 7830-7835
- PubMed: 29797764
- DOI: https://doi.org/10.1002/anie.201803591
- Primary Citation of Related Structures:
5YRT, 5YRV, 5YSH, 5YSN, 5YSR - PubMed Abstract:
The crystal structures of the B 12 -dependent isomerases (eliminating) diol dehydratase and ethanolamine ammonia-lyase complexed with adenosylcobalamin were solved with and without substrates. The structures revealed that the peripheral a-acetamide side chain of the corrin ring directly interacts with the adenosyl group to maintain the group in the catalytic position, and that this side chain swings between the original and catalytic positions in a synchronized manner with the radical shuttling between the coenzyme and substrate/product. Mutations involving key residues that cooperatively participate in the positioning of the adenosyl group, directly or indirectly through the interaction with the a-side chain, decreased the turnover rate and increased the relative rate of irreversible inactivation caused by undesirable side reactions. These findings guide the engineering of enzymes for improved catalysis and producing useful chemicals by utilizing the high reactivity of radical species.
- Department of Picobiology/Life Science, Graduate School of Life Science, University of Hyogo, 3-2-1 Koto, Kamigori-cho, Ako-gun, Hyogo, 678-1297, Japan.
Organizational Affiliation: