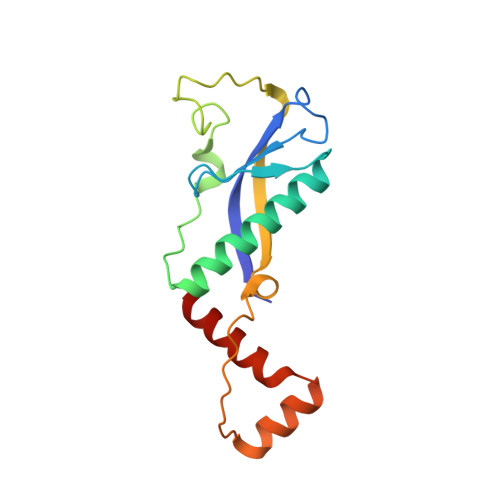
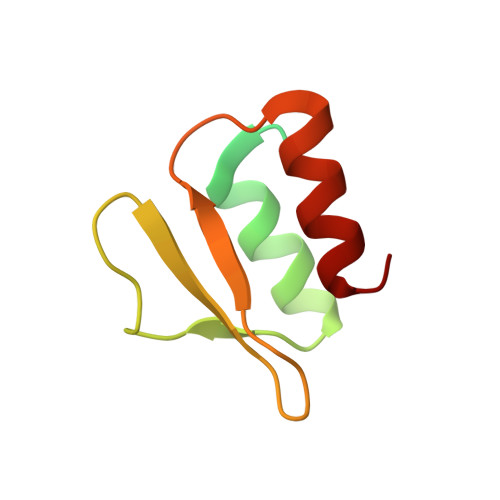
Functional insights into the Streptococcus pneumoniae HicBA toxin-antitoxin system based on a structural study.
Kim, D.H., Kang, S.M., Park, S.J., Jin, C., Yoon, H.J., Lee, B.J.(2018) Nucleic Acids Res 46: 6371-6386
- PubMed: 29878152
- DOI: https://doi.org/10.1093/nar/gky469
- Primary Citation of Related Structures:
5YRZ - PubMed Abstract:
Streptococcus pneumonia has attracted increasing attention due to its resistance to existing antibiotics. TA systems are essential for bacterial persistence under stressful conditions such as nutrient deprivation, antibiotic treatment, and immune system attacks. In particular, S. pneumoniae expresses the HicBA TA gene, which encodes the stable HicA toxin and the labile HicB antitoxin. These proteins interact to form a non-toxic TA complex under normal conditions, but the toxin is activated by release from the antitoxin in response to unfavorable growth conditions. Here, we present the first crystal structure showing the complete conformation of the HicBA complex from S. pneumonia. The structure reveals that the HicA toxin contains a double-stranded RNA-binding domain that is essential for RNA recognition and that the C-terminus of the HicB antitoxin folds into a ribbon-helix-helix DNA-binding motif. The active site of HicA is sterically blocked by the N-terminal region of HicB. RNase activity assays show that His36 is essential for the ribonuclease activity of HicA, and nuclear magnetic resonance (NMR) spectra show that several residues of HicB participate in binding to the promoter DNA of the HicBA operon. A toxin-mimicking peptide that inhibits TA complex formation and thereby increases toxin activity was designed, providing a novel approach to the development of new antibiotics.
- The Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Gwanak-gu, Seoul 08826, Republic of Korea.
Organizational Affiliation: