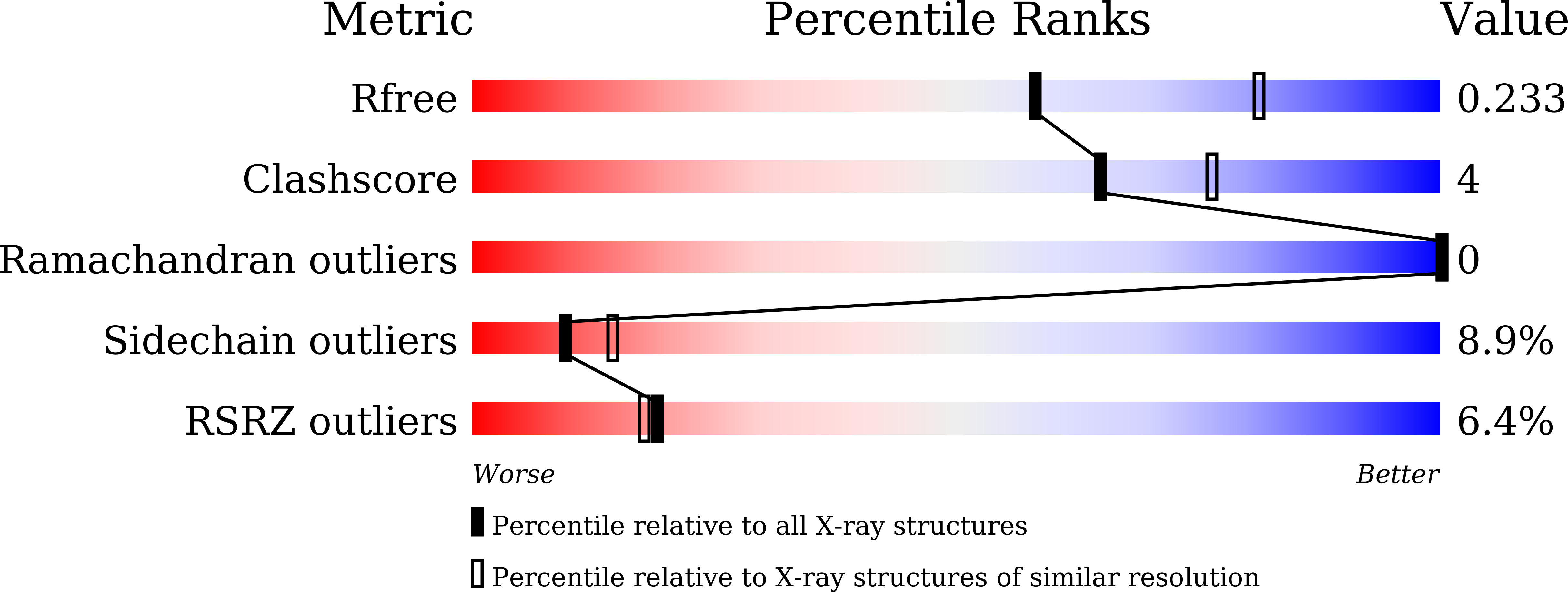
Crystal structure of LysK, an enzyme catalyzing the last step of lysine biosynthesis in Thermus thermophilus, in complex with lysine: Insight into the mechanism for recognition of the amino-group carrier protein, LysW
Fujita, S., Cho, S.-H., Yoshida, A., Hasebe, F., Tomita, T., Kuzuyama, T., Nishiyama, M.(2017) Biochem Biophys Res Commun 491: 409-415
- PubMed: 28720495
- DOI: https://doi.org/10.1016/j.bbrc.2017.07.088
- Primary Citation of Related Structures:
5XOY - PubMed Abstract:
LysK is an M20 peptidase family enzyme that hydrolyzes the isopeptide bond between the carrier protein LysW and lysine in order to release lysine, which is the last step of lysine biosynthesis in Thermus thermophilus. In the present study, we determined the crystal structure of LysK in complex with lysine at a resolution of 2.4 Å. The α-amino group of the bound lysine was oriented toward the catalytic center, which was composed of the residues coordinating divalent metal ions for the hydrolysis of the isopeptide bond. An 11 Å-long path was observed from the active site binding lysine to the protein surface, which may be responsible for recognizing the C-terminal extension domain of LysW with the conserved EDWGE sequence. A positively-charged surface region was detected around the exit of the path, similar to other lysine biosynthetic enzymes using LysW as the carrier protein. Mutational studies of the surface residues provided a plausible model for the electrostatic interaction with LysW.
Organizational Affiliation:
Biotechnology Research Center, The University of Tokyo, Japan.