High Resolution Crystal Structure of Bacillus Licheniformis Gamma Glutamyl Transpeptidase with Acivicin
Kumari, S., Goel, M., Gupta, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma glutamyl transpeptidase | 398 | Bacillus licheniformis | Mutation(s): 0 EC: 2.3.2.2 (PDB Primary Data), 3.4.19.13 (UniProt) | 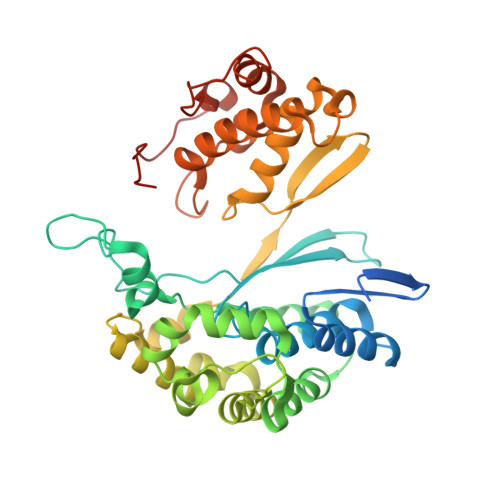 | |
UniProt | |||||
Find proteins for Q65KZ6 (Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46)) Explore Q65KZ6 Go to UniProtKB: Q65KZ6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q65KZ6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma glutamyl transpeptidase | 187 | Bacillus licheniformis | Mutation(s): 0 EC: 2.3.2.2 (PDB Primary Data), 3.4.19.13 (UniProt) | 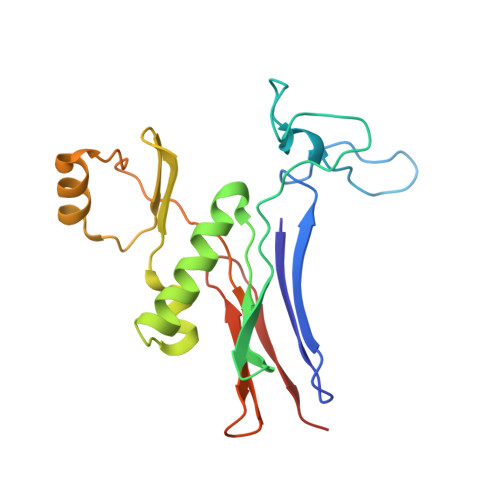 | |
UniProt | |||||
Find proteins for A9YTT0 (Bacillus licheniformis) Explore A9YTT0 Go to UniProtKB: A9YTT0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9YTT0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AVN Query on AVN | D [auth B] | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID C5 H7 Cl N2 O3 QAWIHIJWNYOLBE-OKKQSCSOSA-N |  | ||
| GOL Query on GOL | C [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CA Query on CA | E [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.005 | α = 90 |
| b = 85.732 | β = 90 |
| c = 124.664 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data processing |
| PHASER | model building |