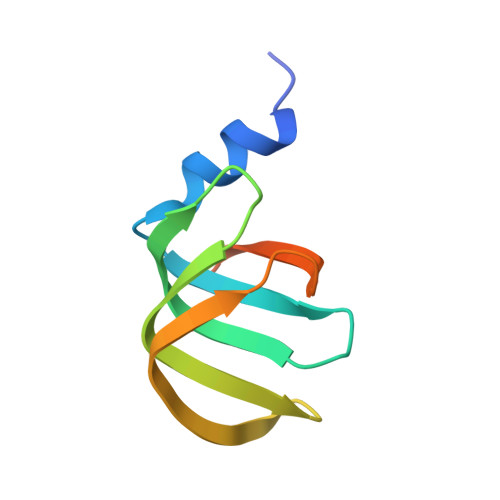
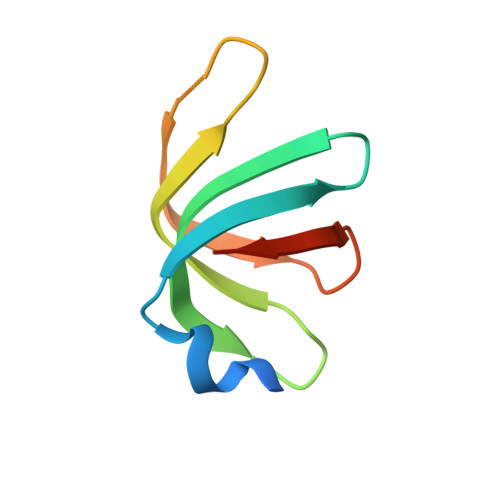
Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly
Zhang, R., So, B.R., Li, P., Yong, J., Glisovic, T., Wan, L., Dreyfuss, G.(2011) Cell 146: 384-395
- PubMed: 21816274
- DOI: https://doi.org/10.1016/j.cell.2011.06.043
- Primary Citation of Related Structures:
5XJL - PubMed Abstract:
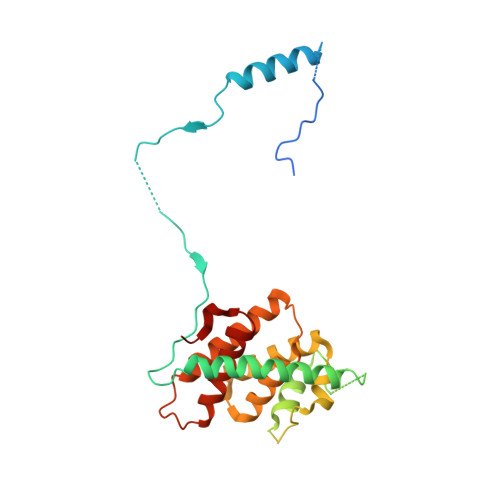
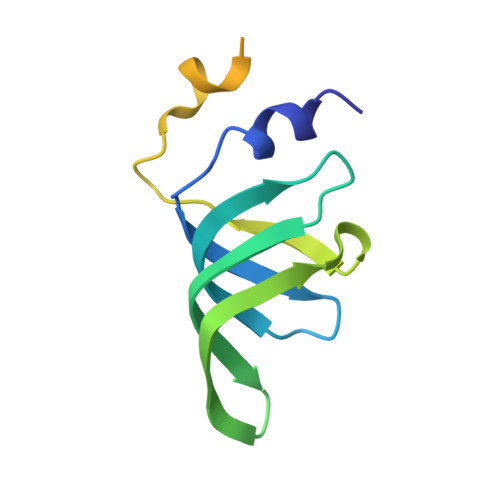
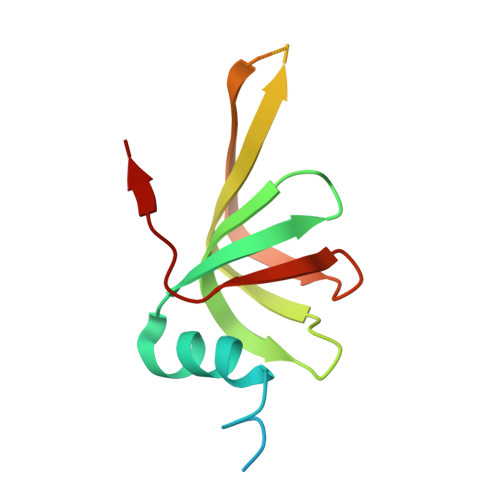
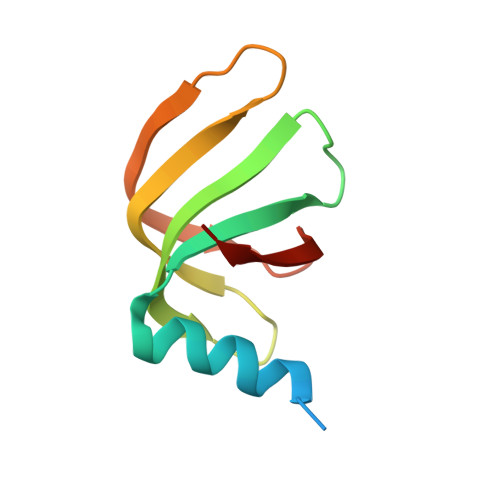
The SMN complex mediates the assembly of heptameric Sm protein rings on small nuclear RNAs (snRNAs), which are essential for snRNP function. Specific Sm core assembly depends on Sm proteins and snRNA recognition by SMN/Gemin2- and Gemin5-containing subunits, respectively. The mechanism by which the Sm proteins are gathered while preventing illicit Sm assembly on non-snRNAs is unknown. Here, we describe the 2.5 Å crystal structure of Gemin2 bound to SmD1/D2/F/E/G pentamer and SMN's Gemin2-binding domain, a key assembly intermediate. Remarkably, through its extended conformation, Gemin2 wraps around the crescent-shaped pentamer, interacting with all five Sm proteins, and gripping its bottom and top sides and outer perimeter. Gemin2 reaches into the RNA-binding pocket, preventing RNA binding. Interestingly, SMN-Gemin2 interaction is abrogated by a spinal muscular atrophy (SMA)-causing mutation in an SMN helix that mediates Gemin2 binding. These findings provide insight into SMN complex assembly and specificity, linking snRNP biogenesis and SMA pathogenesis.
- Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, PA 19104-6148, USA.
Organizational Affiliation: