A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Wang, D., Chen, C., Liu, S., Zhou, H., Yang, K., Zhao, Q., Ji, X., Chen, C., Xie, W., Wang, Z., Mi, L.Z., Yang, H.(2017) Sci Rep 7: 42580-42580
- PubMed: 28198446
- DOI: https://doi.org/10.1038/srep42580
- Primary Citation of Related Structures:
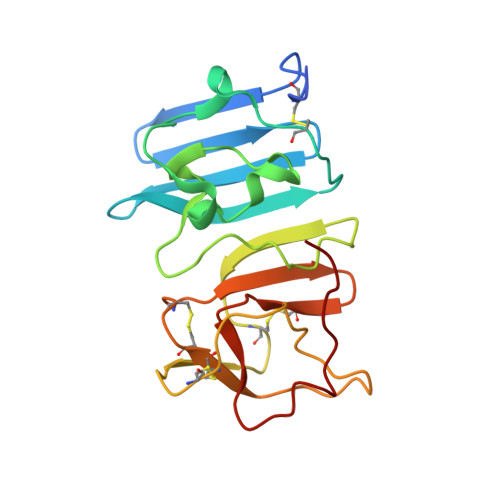
5X8Y - PubMed Abstract:
An unprecedented epidemic of Zika virus (ZIKV) infection had spread to South and Central America. ZIKV infection was recently confirmed by CDC (the Centers for Disease Control and Prevention) to cause neonatal microcephaly, which posed a significant public health emergency of international concern. No specific vaccines or drugs are currently available to fight ZIKV infection. ZIKV nonstructural protein 1 (NS1) plays an essential role in viral replication and immune evasion. We determined the crystal structure of ZIKV NS1 172-352 , which forms a head-to-head, symmetric dimer with a unique 14-stranded β-ladder conserved among flaviviruses. The assembly of the β-ladder dimer is concentration dependent. Strikingly, one pathogenic mutation T233A (NCBI accession no. KU527068), found in the brain tissue of infected fetus with neonatal microcephaly, is located at the dimer interface. Thr233, a unique residue found in ZIKV but not in other flaviviruses, organizes a central hydrogen bonding network at NS1 dimer interface. Mutation of Thr233 to Ala disrupts this elaborated interaction network, and destabilizes the NS1 dimeric assembly in vitro. In addition, our structural comparison of epitopes for protective antibody 22NS1, targeting West Nile Virus NS1, could potentially be valuable in understanding its anti-virus specificities and in the development of antibodies against ZIKV.
- School of Life Sciences, Tianjin University, Tianjin 300072, People's Republic of China.
Organizational Affiliation: