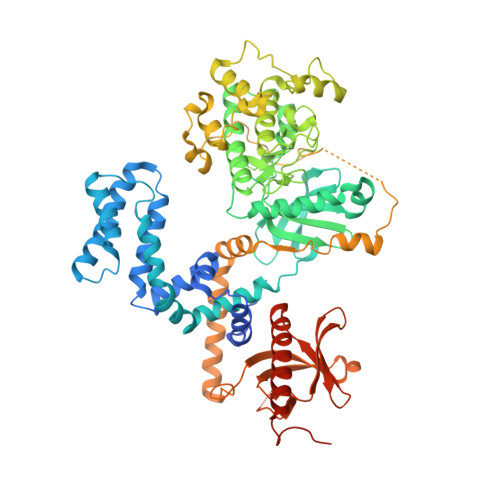
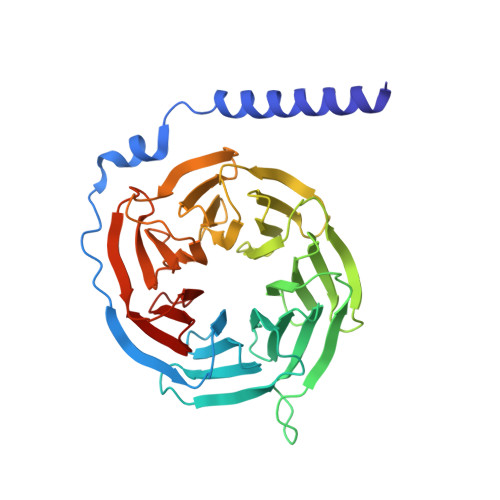
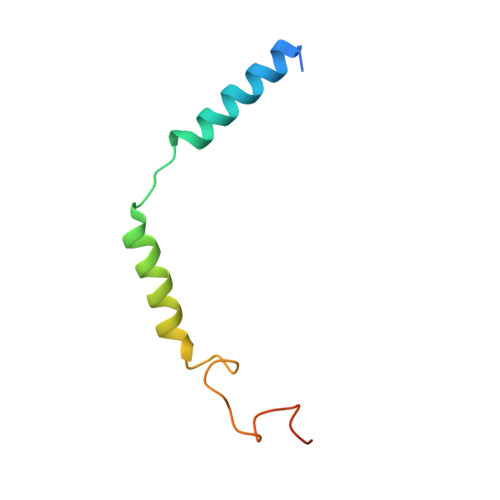
Structural Determinants Influencing the Potency and Selectivity of Indazole-Paroxetine Hybrid G Protein-Coupled Receptor Kinase 2 Inhibitors.
Bouley, R., Waldschmidt, H.V., Cato, M.C., Cannavo, A., Song, J., Cheung, J.Y., Yao, X.Q., Koch, W.J., Larsen, S.D., Tesmer, J.J.G.(2017) Mol Pharmacol 92: 707-717
- PubMed: 29070696
- DOI: https://doi.org/10.1124/mol.117.110130
- Primary Citation of Related Structures:
5WG3, 5WG4, 5WG5 - PubMed Abstract:
G protein-coupled receptor kinases (GRKs) phosphorylate activated receptors to promote arrestin binding, decoupling from heterotrimeric G proteins, and internalization. GRK2 and GRK5 are overexpressed in the failing heart and thus have become therapeutic targets. Previously, we discovered two classes of GRK2-selective inhibitors, one stemming from GSK180736A, a Rho-associated coiled-coil containing kinase 1 (ROCK1) inhibitor, the other from paroxetine, a selective serotonin-reuptake inhibitor. These two classes of compounds bind to the GRK2 active site in a similar configuration but contain different hinge-binding "warheads": indazole and benzodioxole, respectively. We surmised from our prior studies that an indazole would be the stronger hinge binder and would impart increased potency when substituted for benzodioxole in paroxetine derivatives. To test this hypothesis, we synthesized a series of hybrid compounds that allowed us to compare the effects of inhibitors that differ only in the identity of the warhead. The indazole-paroxetine analogs were indeed more potent than their respective benzodioxole derivatives but lost selectivity. To investigate how these two warheads dictate selectivity, we determined the crystal structures of three of the indazole hybrid compounds (CCG224061, CCG257284, and CCG258748) in complex with GRK2-G βγ Comparison of these structures with those of analogous benzodioxole-containing complexes confirmed that the indazole-paroxetine hybrids form stronger interactions with the hinge of the kinase but also stabilize a distinct conformation of the kinase domain of GRK2 compared with previous complexes with paroxetine analogs. This conformation is analogous to one that can be assumed by GRK5, at least partially explaining the loss in selectivity.
- Life Sciences Institute (R.B., H.V.W., M.C.C., J.J.G.T.), Departments of Medicinal Chemistry (H.V.W., S.D.L., J.J.G.T.), Pharmacology (R.B., J.J.G.T.), Biological Chemistry (M.C.C., J.J.G.T.), and Vahlteich Medicinal Chemistry Core, College of Pharmacy (H.V.W., S.D.L.), University of Michigan, Ann Arbor, Michigan; Department of Chemistry, Georgia State University, Atlanta, Georgia (X.-Q.Y.); Center for Translational Medicine, Temple University, Philadelphia, Pennsylvania (A.C., J.S., J.Y.C, W.J.K.); and Department of Biological Sciences, Purdue University, West Lafayette Indiana (J.J.G.T.).
Organizational Affiliation: