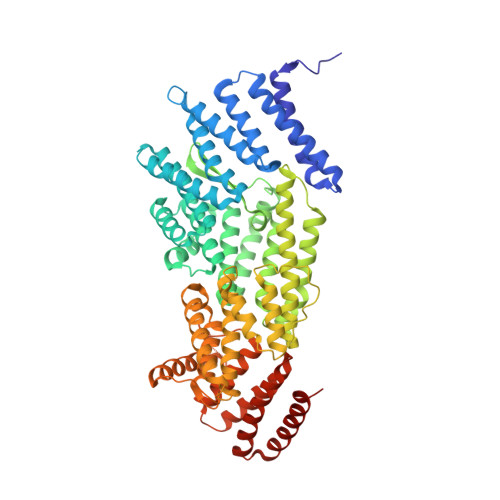
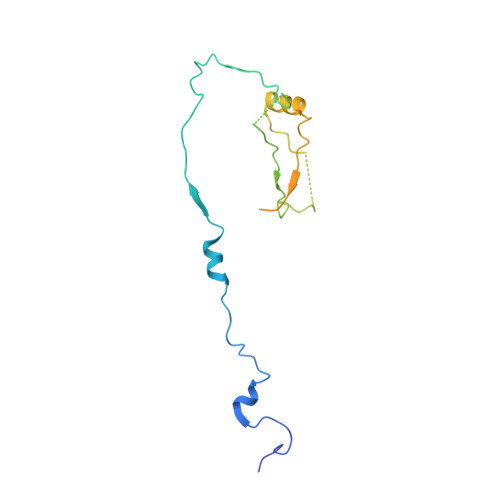
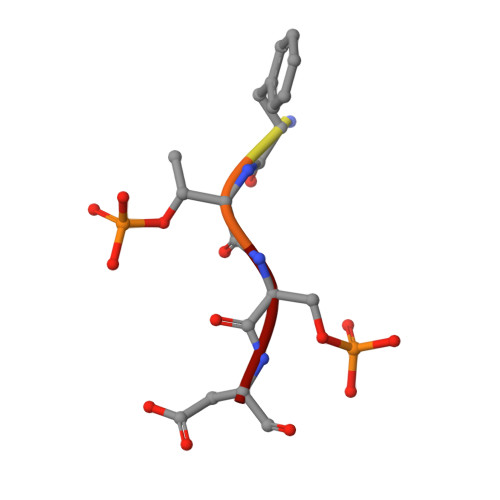
The Kinetochore Receptor for the Cohesin Loading Complex.
Hinshaw, S.M., Makrantoni, V., Harrison, S.C., Marston, A.L.(2017) Cell 171: 72-84.e13
- PubMed: 28938124
- DOI: https://doi.org/10.1016/j.cell.2017.08.017
- Primary Citation of Related Structures:
5W94 - PubMed Abstract:
The ring-shaped cohesin complex brings together distant DNA domains to maintain, express, and segregate the genome. Establishing specific chromosomal linkages depends on cohesin recruitment to defined loci. One such locus is the budding yeast centromere, which is a paradigm for targeted cohesin loading. The kinetochore, a multiprotein complex that connects centromeres to microtubules, drives the recruitment of high levels of cohesin to link sister chromatids together. We have exploited this system to determine the mechanism of specific cohesin recruitment. We show that phosphorylation of the Ctf19 kinetochore protein by a conserved kinase, DDK, provides a binding site for the Scc2/4 cohesin loading complex, thereby directing cohesin loading to centromeres. A similar mechanism targets cohesin to chromosomes in vertebrates. These findings represent a complete molecular description of targeted cohesin loading, a phenomenon with wide-ranging importance in chromosome segregation and, in multicellular organisms, transcription regulation.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: