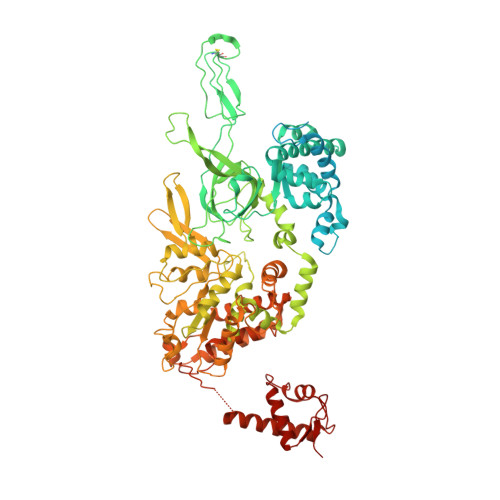
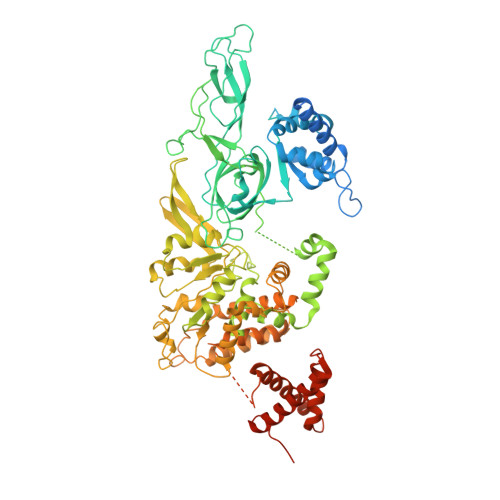
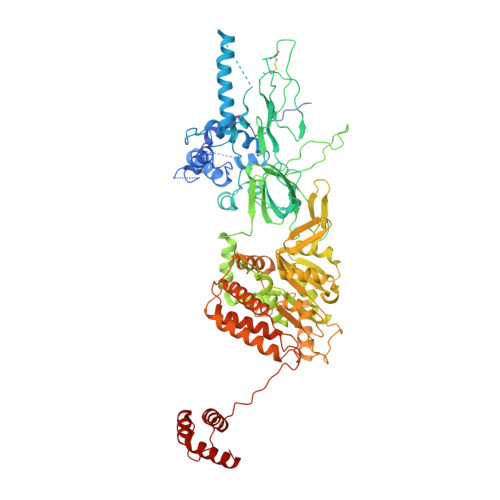
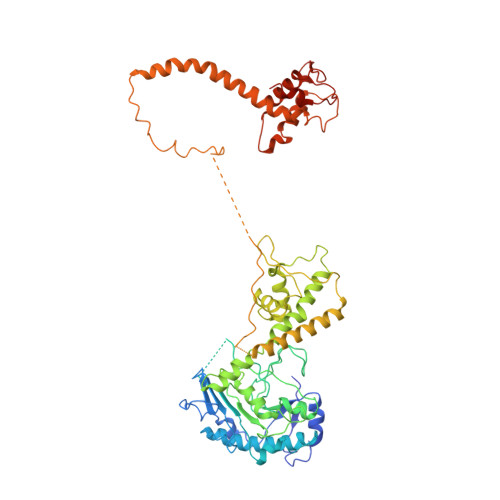
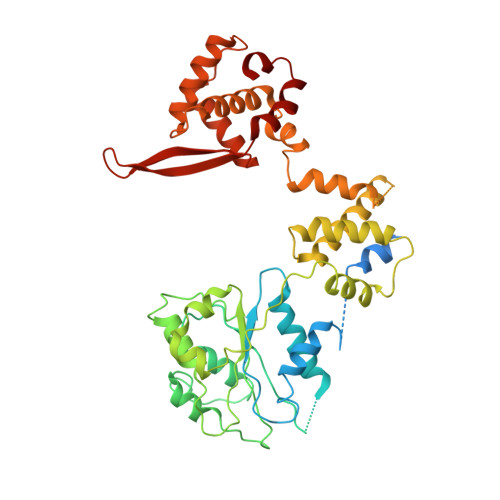
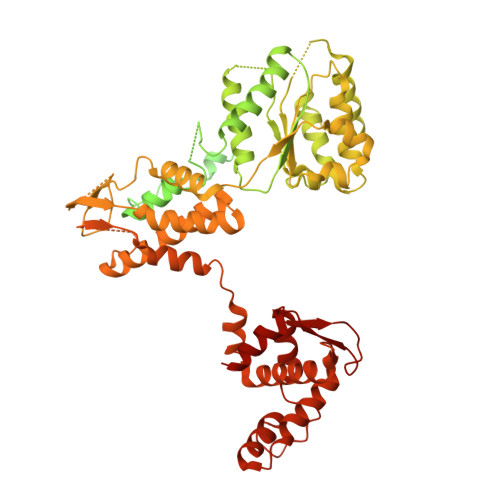
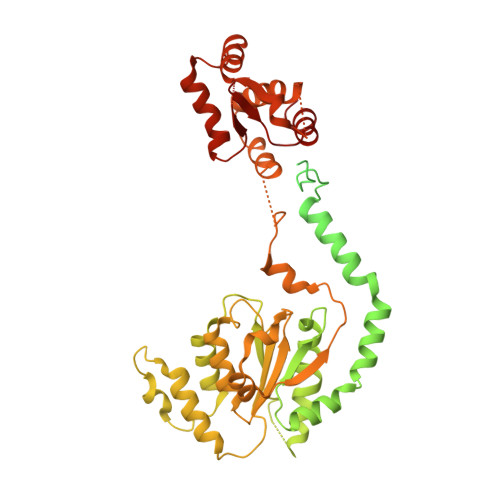
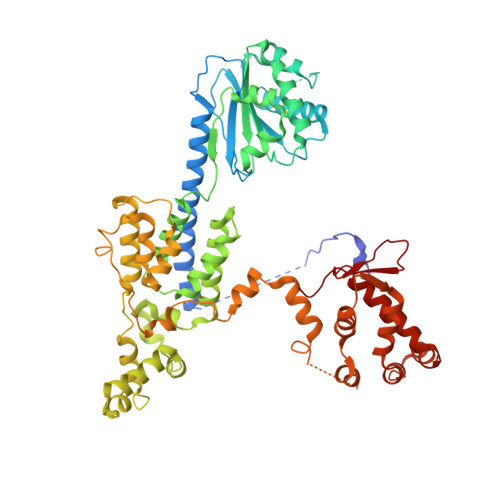
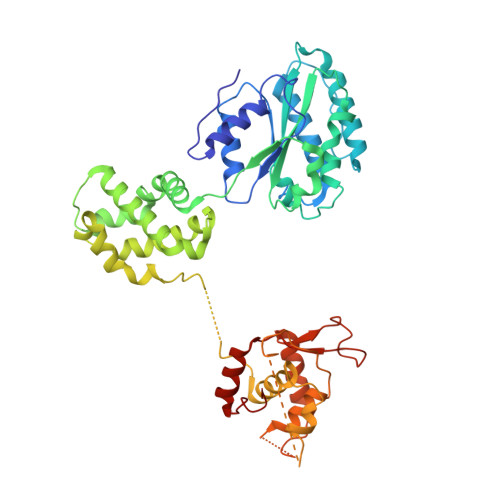
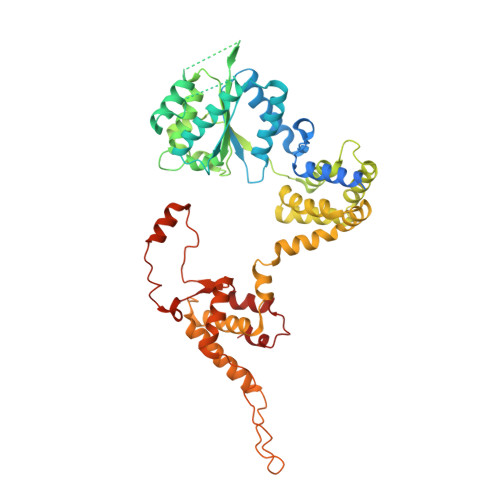
Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Yuan, Z., Riera, A., Bai, L., Sun, J., Nandi, S., Spanos, C., Chen, Z.A., Barbon, M., Rappsilber, J., Stillman, B., Speck, C., Li, H.(2017) Nat Struct Mol Biol 24: 316-324
- PubMed: 28191893
- DOI: https://doi.org/10.1038/nsmb.3372
- Primary Citation of Related Structures:
5V8F - PubMed Abstract:
To initiate DNA replication, the origin recognition complex (ORC) and Cdc6 load an Mcm2-7 double hexamer onto DNA. Without ATP hydrolysis, ORC-Cdc6 recruits one Cdt1-bound Mcm2-7 hexamer, thus forming an ORC-Cdc6-Cdt1-Mcm2-7 (OCCM) helicase-loading intermediate. Here we report a 3.9-Å structure of Saccharomyces cerevisiae OCCM on DNA. Flexible Mcm2-7 winged-helix domains (WHDs) engage ORC-Cdc6. A three-domain Cdt1 configuration embraces Mcm2, Mcm4, and Mcm6, thus comprising nearly half of the hexamer. The Cdt1 C-terminal domain extends to the Mcm6 WHD, which binds the Orc4 WHD. DNA passes through the ORC-Cdc6 and Mcm2-7 rings. Origin DNA interaction is mediated by an α-helix within Orc4 and positively charged loops within Orc2 and Cdc6. The Mcm2-7 C-tier AAA+ ring is topologically closed by an Mcm5 loop that embraces Mcm2, but the N-tier-ring Mcm2-Mcm5 interface remains open. This structure suggests a loading mechanism of the first Cdt1-bound Mcm2-7 hexamer by ORC-Cdc6.
- Cryo-EM Structural Biology Laboratory, Van Andel Research Institute, Grand Rapids, Michigan, USA.
Organizational Affiliation: