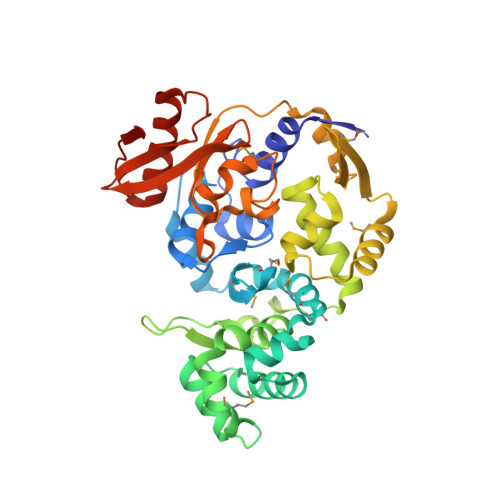
Structure of a DNA glycosylase that unhooks interstrand cross-links.
Mullins, E.A., Warren, G.M., Bradley, N.P., Eichman, B.F.(2017) Proc Natl Acad Sci U S A 114: 4400-4405
- PubMed: 28396405
- DOI: https://doi.org/10.1073/pnas.1703066114
- Primary Citation of Related Structures:
5UUJ - PubMed Abstract:
DNA glycosylases are important editing enzymes that protect genomic stability by excising chemically modified nucleobases that alter normal DNA metabolism. These enzymes have been known only to initiate base excision repair of small adducts by extrusion from the DNA helix. However, recent reports have described both vertebrate and microbial DNA glycosylases capable of unhooking highly toxic interstrand cross-links (ICLs) and bulky minor groove adducts normally recognized by Fanconi anemia and nucleotide excision repair machinery, although the mechanisms of these activities are unknown. Here we report the crystal structure of Streptomyces sahachiroi AlkZ (previously Orf1), a bacterial DNA glycosylase that protects its host by excising ICLs derived from azinomycin B (AZB), a potent antimicrobial and antitumor genotoxin. AlkZ adopts a unique fold in which three tandem winged helix-turn-helix motifs scaffold a positively charged concave surface perfectly shaped for duplex DNA. Through mutational analysis, we identified two glutamine residues and a β-hairpin within this putative DNA-binding cleft that are essential for catalytic activity. Additionally, we present a molecular docking model for how this active site can unhook either or both sides of an AZB ICL, providing a basis for understanding the mechanisms of base excision repair of ICLs. Given the prevalence of this protein fold in pathogenic bacteria, this work also lays the foundation for an emerging role of DNA repair in bacteria-host pathogenesis.
- Department of Biological Sciences, Vanderbilt University, Nashville, TN 37232.
Organizational Affiliation: