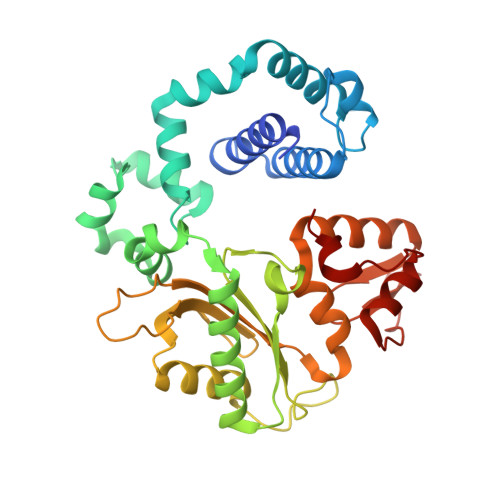
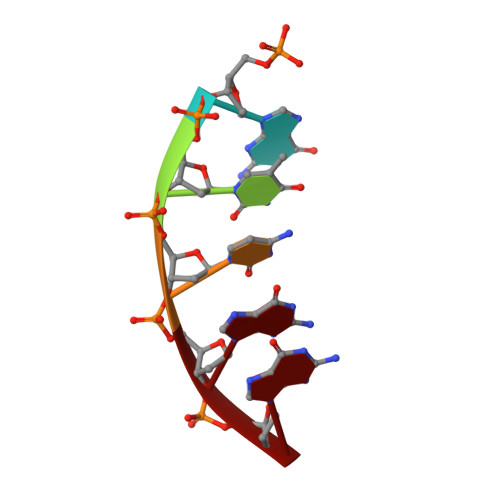
Modulating the DNA polymerase beta reaction equilibrium to dissect the reverse reaction.
Shock, D.D., Freudenthal, B.D., Beard, W.A., Wilson, S.H.(2017) Nat Chem Biol 13: 1074-1080
- PubMed: 28759020
- DOI: https://doi.org/10.1038/nchembio.2450
- Primary Citation of Related Structures:
5UGN, 5UGO, 5UGP - PubMed Abstract:
DNA polymerases catalyze efficient and high-fidelity DNA synthesis. While this reaction favors nucleotide incorporation, polymerases also catalyze a reverse reaction, pyrophosphorolysis, that removes the DNA primer terminus and generates deoxynucleoside triphosphates. Because pyrophosphorolysis can influence polymerase fidelity and sensitivity to chain-terminating nucleosides, we analyzed pyrophosphorolysis with human DNA polymerase β and found the reaction to be inefficient. The lack of a thio-elemental effect indicated that this reaction was limited by a nonchemical step. Use of a pyrophosphate analog, in which the bridging oxygen is replaced with an imido group (PNP), increased the rate of the reverse reaction and displayed a large thio-elemental effect, indicating that chemistry was now rate determining. Time-lapse crystallography with PNP captured structures consistent with a chemical equilibrium favoring the reverse reaction. These results highlight the importance of the bridging atom between the β- and γ-phosphates of the incoming nucleotide in reaction chemistry, enzyme conformational changes, and overall reaction equilibrium.
- Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, North Carolina, USA.
Organizational Affiliation: