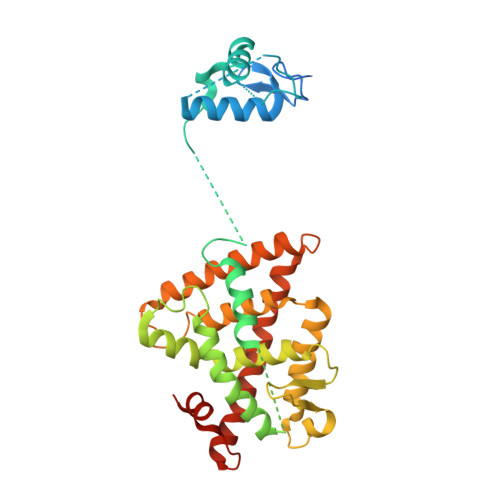
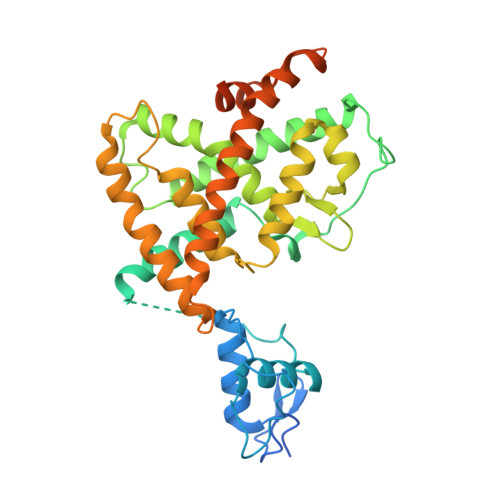
The quaternary architecture of RAR beta-RXR alpha heterodimer facilitates domain-domain signal transmission.
Chandra, V., Wu, D., Li, S., Potluri, N., Kim, Y., Rastinejad, F.(2017) Nat Commun 8: 868-868
- PubMed: 29021580
- DOI: https://doi.org/10.1038/s41467-017-00981-y
- Primary Citation of Related Structures:
5UAN - PubMed Abstract:
Assessing the physical connections and allosteric communications in multi-domain nuclear receptor (NR) polypeptides has remained challenging, with few crystal structures available to show their overall structural organizations. Here we report the quaternary architecture of multi-domain retinoic acid receptor β-retinoic X receptor α (RARβ-RXRα) heterodimer bound to DNA, ligands and coactivator peptides, examined through crystallographic, hydrogen-deuterium exchange mass spectrometry, mutagenesis and functional studies. The RARβ ligand-binding domain (LBD) and DNA-binding domain (DBD) are physically connected to foster allosteric signal transmission between them. Direct comparisons among all the multi-domain NRs studied crystallographically to date show significant variations within their quaternary architectures, rather than a common architecture adhering to strict rules. RXR remains flexible and adaptive by maintaining loosely organized domains, while its heterodimerization partners use a surface patch on their LBDs to form domain-domain interactions with DBDs.Nuclear receptors (NR) are multidomain proteins, which makes their crystallization challenging. Here the authors present the crystal structure of the retinoic acid receptor β-retinoic X receptor α (RARβ-RXRα) heterodimer bound to DNA, ligands and coactivator peptides, which shows that NR quaternary architectures are variable.
- Integrative Metabolism Program, Sanford Burnham Prebys Medical Discovery Institute, Orlando, FL, 32827, USA.
Organizational Affiliation: