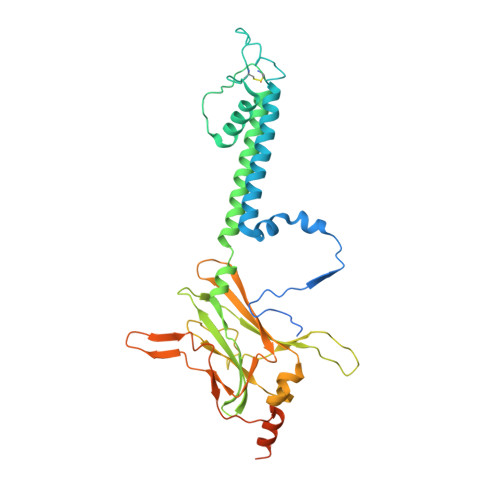
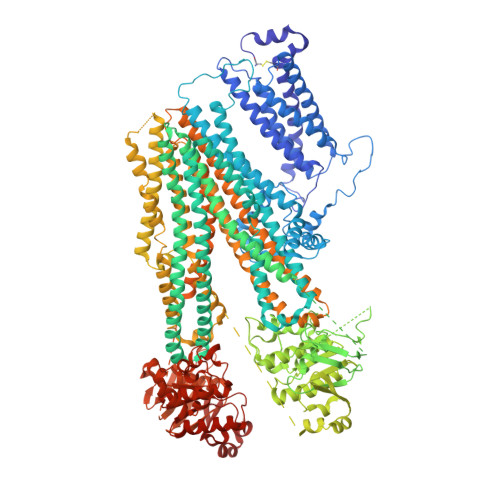
Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Martin, G.M., Yoshioka, C., Rex, E.A., Fay, J.F., Xie, Q., Whorton, M.R., Chen, J.Z., Shyng, S.L.(2017) Elife 6
- PubMed: 28092267
- DOI: https://doi.org/10.7554/eLife.24149
- Primary Citation of Related Structures:
5TWV - PubMed Abstract:
K ATP channels are metabolic sensors that couple cell energetics to membrane excitability. In pancreatic β-cells, channels formed by SUR1 and Kir6.2 regulate insulin secretion and are the targets of antidiabetic sulfonylureas. Here, we used cryo-EM to elucidate structural basis of channel assembly and gating. The structure, determined in the presence of ATP and the sulfonylurea glibenclamide, at ~6 Å resolution reveals a closed Kir6.2 tetrameric core with four peripheral SUR1s each anchored to a Kir6.2 by its N-terminal transmembrane domain (TMD0). Intricate interactions between TMD0, the loop following TMD0, and Kir6.2 near the proposed PIP 2 binding site, and where ATP density is observed, suggest SUR1 may contribute to ATP and PIP 2 binding to enhance Kir6.2 sensitivity to both. The SUR1-ABC core is found in an unusual inward-facing conformation whereby the two nucleotide binding domains are misaligned along a two-fold symmetry axis, revealing a possible mechanism by which glibenclamide inhibits channel activity.
- Department of Biochemistry and Molecular Biology, Oregon Health and Science University, Portland, Oregon, United States.
Organizational Affiliation: