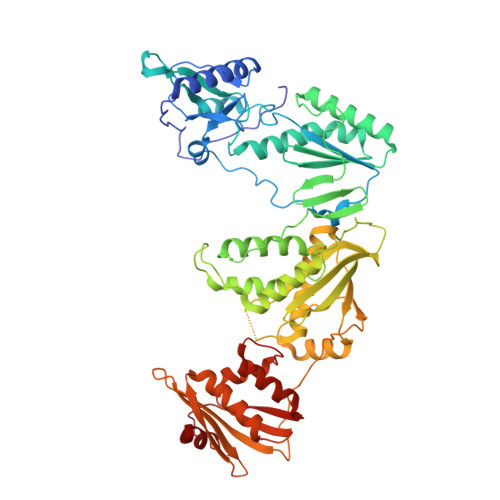
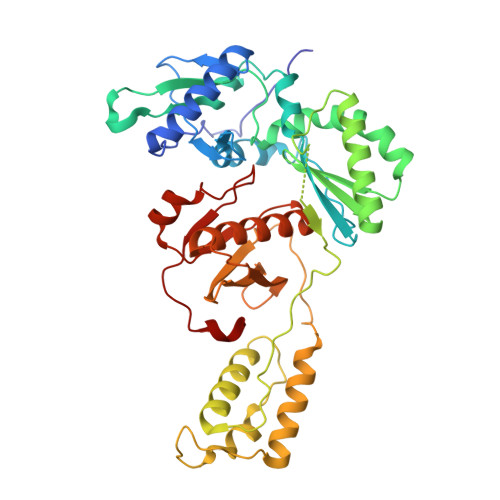
Structural and Preclinical Studies of Computationally Designed Non-Nucleoside Reverse Transcriptase Inhibitors for Treating HIV infection.
Kudalkar, S.N., Beloor, J., Chan, A.H., Lee, W.G., Jorgensen, W.L., Kumar, P., Anderson, K.S.(2017) Mol Pharmacol 91: 383-391
- PubMed: 28167742
- DOI: https://doi.org/10.1124/mol.116.107755
- Primary Citation of Related Structures:
5TW3 - PubMed Abstract:
The clinical benefits of HIV-1 non-nucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are hindered by their unsatisfactory pharmacokinetic (PK) properties along with the rapid development of drug-resistant variants. However, the clinical efficacy of these inhibitors can be improved by developing compounds with enhanced pharmacological profiles and heightened antiviral activity. We used computational and structure-guided design to develop two next-generation NNRTI drug candidates, compounds I and II, which are members of a class of catechol diethers. We evaluated the preclinical potential of these compounds in BALB/c mice because of their high solubility (510 µ g/ml for compound I and 82.9 µ g/ml for compound II), low cytotoxicity, and enhanced antiviral activity against wild-type (WT) HIV-1 RT and resistant variants. Additionally, crystal structures of compounds I and II with WT RT suggested an optimal binding to the NNRTI binding pocket favoring the high anti-viral potency. A single intraperitoneal dose of compounds I and II exhibited a prolonged serum residence time of 48 hours and concentration maximum ( C max ) of 4000- to 15,000-fold higher than their therapeutic/effective concentrations. These C max values were 4- to 15-fold lower than their cytotoxic concentrations observed in MT-2 cells. Compound II showed an enhanced area under the curve (0-last) and decreased plasma clearance over compound I and efavirenz, the standard of care NNRTI. Hence, the overall (PK) profile of compound II was excellent compared with that of compound I and efavirenz. Furthermore, both compounds were very well tolerated in BALB/c mice without any detectable acute toxicity. Taken together, these data suggest that compounds I and II possess improved anti-HIV-1 potency, remarkable in vivo safety, and prolonged in vivo circulation time, suggesting strong potential for further development as new NNRTIs for the potential treatment of HIV infection.
- Departments of Pharmacology, School of Medicine (S.N.K., A.H.C., K.S.A.), Infectious Diseases/Internal Medicine, School of Medicine (J.B., P.K.), and Chemistry (W.-G.L., W.L.J.), Yale University, New Haven, Connecticut.
Organizational Affiliation: