Structural insights into NusG regulating transcription elongation.
Liu, B., Steitz, T.A.(2017) Nucleic Acids Res 45: 968-974
- PubMed: 27899640
- DOI: https://doi.org/10.1093/nar/gkw1159
- Primary Citation of Related Structures:
5TBZ - PubMed Abstract:
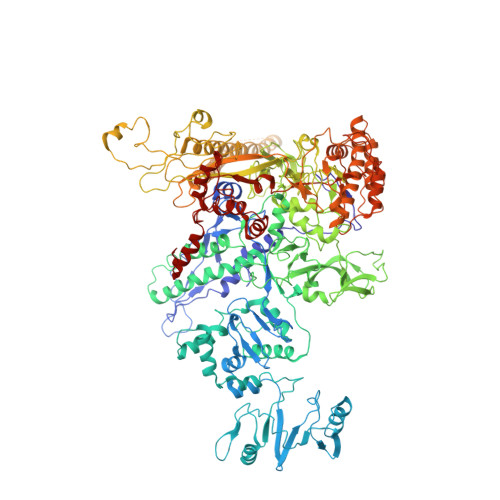
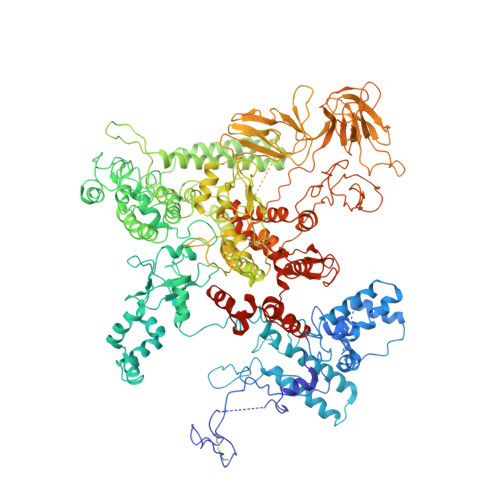
NusG is an essential transcription factor that plays multiple key regulatory roles in transcription elongation, termination and coupling translation and transcription. The core role of NusG is to enhance transcription elongation and RNA polymerase processivity. Here, we present the structure of Escherichia coli RNA polymerase complexed with NusG. The structure shows that the NusG N-terminal domain (NGN) binds at the central cleft of RNA polymerase surrounded by the β' clamp helices, the β protrusion, and the β lobe domains to close the promoter DNA binding channel and constrain the β' clamp domain, but with an orientation that is different from the one observed in the archaeal β' clamp-Spt4/5 complex. The structure also allows us to construct a reliable model of the complete NusG-associated transcription elongation complex, suggesting that the NGN domain binds at the upstream fork junction of the transcription elongation complex, similar to σ2 in the transcription initiation complex, to stabilize the junction, and therefore enhances transcription processivity.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: