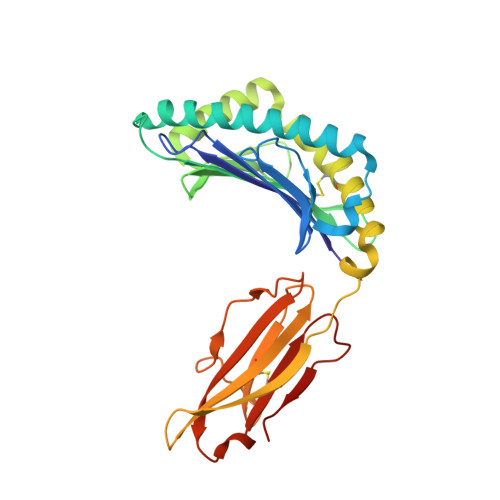
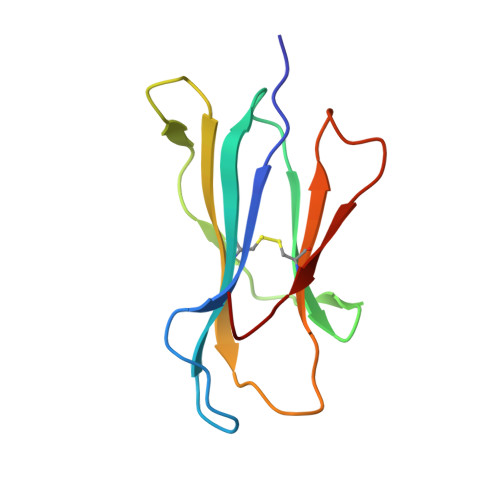
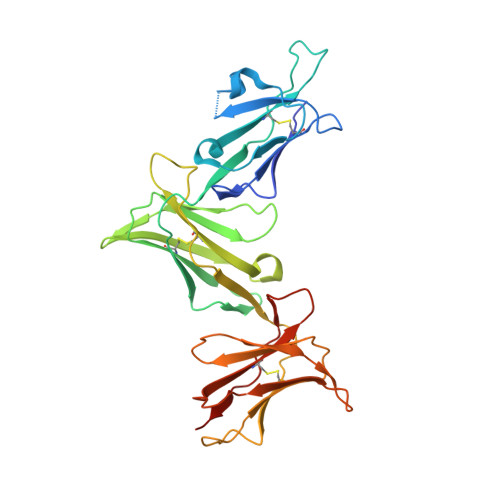
MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Pymm, P., Illing, P.T., Ramarathinam, S.H., O'Connor, G.M., Hughes, V.A., Hitchen, C., Price, D.A., Ho, B.K., McVicar, D.W., Brooks, A.G., Purcell, A.W., Rossjohn, J., Vivian, J.P.(2017) Nat Struct Mol Biol 24: 387-394
- PubMed: 28218747
- DOI: https://doi.org/10.1038/nsmb.3381
- Primary Citation of Related Structures:
5T6W, 5T6X, 5T6Y, 5T6Z, 5T70 - PubMed Abstract:
Major histocompatibility complex class I (MHC-I) molecules play a crucial role in immunity by capturing peptides for presentation to T cells and natural killer (NK) cells. The peptide termini are tethered within the MHC-I antigen-binding groove, but it is unknown whether other presentation modes occur. Here we show that 20% of the HLA-B*57:01 peptide repertoire comprises N-terminally extended sets characterized by a common motif at position 1 (P1) to P2. Structures of HLA-B*57:01 presenting N-terminally extended peptides, including the immunodominant HIV-1 Gag epitope TW10 (TSTLQEQIGW), showed that the N terminus protrudes from the peptide-binding groove. The common escape mutant TSNLQEQIGW bound HLA-B*57:01 canonically, adopting a dramatically different conformation than the TW10 peptide. This affected recognition by killer cell immunoglobulin-like receptor (KIR) 3DL1 expressed on NK cells. We thus define a previously uncharacterized feature of the human leukocyte antigen class I (HLA-I) immunopeptidome that has implications for viral immune escape. We further suggest that recognition of the HLA-B*57:01-TW10 epitope is governed by a 'molecular tension' between the adaptive and innate immune systems.
- Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: