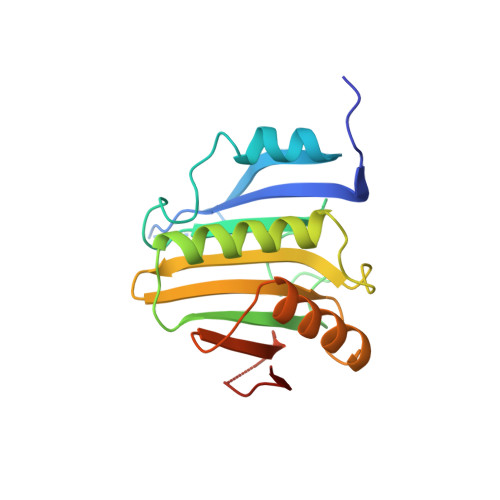
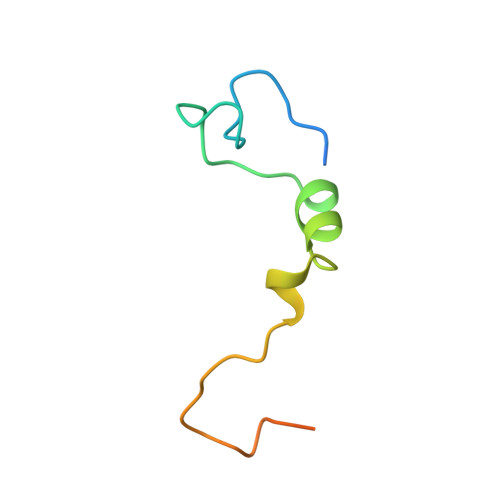
The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Gruner, S., Peter, D., Weber, R., Wohlbold, L., Chung, M.Y., Weichenrieder, O., Valkov, E., Igreja, C., Izaurralde, E.(2016) Mol Cell 64: 467-479
- PubMed: 27773676
- DOI: https://doi.org/10.1016/j.molcel.2016.09.020
- Primary Citation of Related Structures:
5T46, 5T47, 5T48 - PubMed Abstract:
Eukaryotic initiation factor 4G (eIF4G) plays a central role in translation initiation through its interactions with the cap-binding protein eIF4E. This interaction is a major drug target for repressing translation and is naturally regulated by 4E-binding proteins (4E-BPs). 4E-BPs and eIF4G compete for binding to the eIF4E dorsal surface via a shared canonical 4E-binding motif, but also contain auxiliary eIF4E-binding sequences, which were assumed to contact non-overlapping eIF4E surfaces. However, it is unknown how metazoan eIF4G auxiliary sequences bind eIF4E. Here, we describe crystal structures of human and Drosophila melanogaster eIF4E-eIF4G complexes, which unexpectedly reveal that the eIF4G auxiliary sequences bind to the lateral surface of eIF4E, using a similar mode to that of 4E-BPs. Our studies provide a molecular model of the eIF4E-eIF4G complex, shed light on the competition mechanism of 4E-BPs, and enable the rational design of selective eIF4G inhibitors to dampen dysregulated translation in disease.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Spemannstrasse 35, 72076 Tübingen, Germany.
Organizational Affiliation: