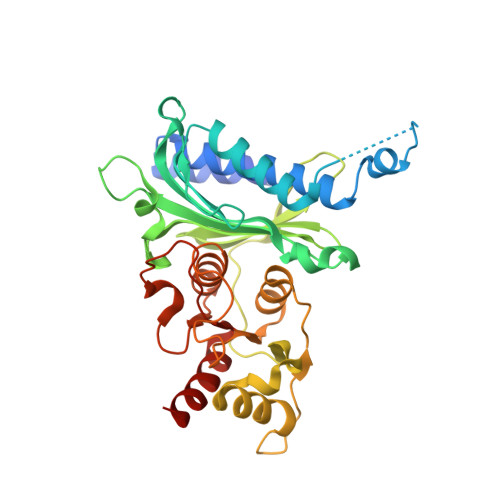
Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(3,4-dichlorophenyl)sulfonylurea
Ruf, A., Joseph, C., Alker, A., Banner, D., Tetaz, T., Benz, J., Kuhn, B., Rudolph, M.G.To be published.