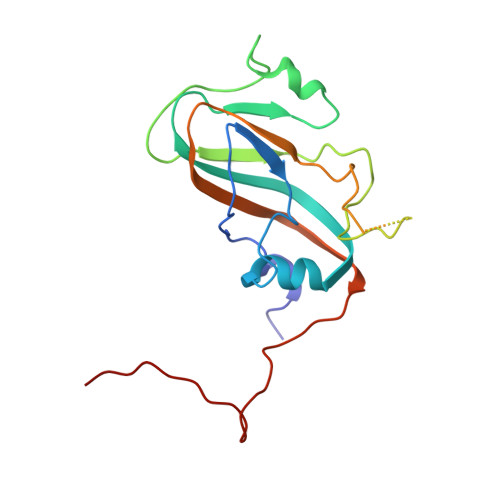
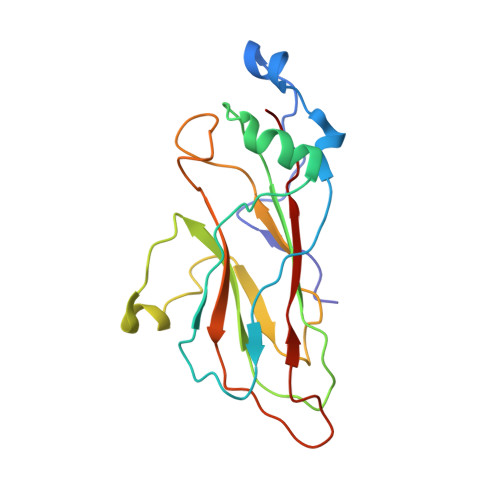
Structures of foot and mouth disease virus pentamers: Insight into capsid dissociation and unexpected pentamer reassociation.
Malik, N., Kotecha, A., Gold, S., Asfor, A., Ren, J., Huiskonen, J.T., Tuthill, T.J., Fry, E.E., Stuart, D.I.(2017) PLoS Pathog 13: e1006607-e1006607
- PubMed: 28937999
- DOI: https://doi.org/10.1371/journal.ppat.1006607
- Primary Citation of Related Structures:
5OWX, 5OYI - PubMed Abstract:
Foot-and-mouth disease virus (FMDV) belongs to the Aphthovirus genus of the Picornaviridae, a family of small, icosahedral, non-enveloped, single-stranded RNA viruses. It is a highly infectious pathogen and is one of the biggest hindrances to the international trade of animals and animal products. FMDV capsids (which are unstable below pH6.5) release their genome into the host cell from an acidic compartment, such as that of an endosome, and in the process dissociate into pentamers. Whilst other members of the family (enteroviruses) have been visualized to form an expanded intermediate capsid with holes from which inner capsid proteins (VP4), N-termini (VP1) and RNA can be released, there has been no visualization of any such state for an aphthovirus, instead the capsid appears to simply dissociate into pentamers. Here we present the 8-Å resolution structure of isolated dissociated pentamers of FMDV, lacking VP4. We also found these pentamers to re-associate into a rigid, icosahedrally symmetric assembly, which enabled their structure to be solved at higher resolution (5.2 Å). In this assembly, the pentamers unexpectedly associate 'inside out', but still with their exposed hydrophobic edges buried. Stabilizing interactions occur between the HI loop of VP2 and its symmetry related partners at the icosahedral 3-fold axes, and between the BC and EF loops of VP3 with the VP2 βB-strand and the CD loop at the 2-fold axes. A relatively extensive but subtle structural rearrangement towards the periphery of the dissociated pentamer compared to that in the mature virus provides insight into the mechanism of dissociation of FMDV and the marked difference in antigenicity.
- Division of Structural Biology, University of Oxford, Headington, Oxford, United Kingdom.
Organizational Affiliation: