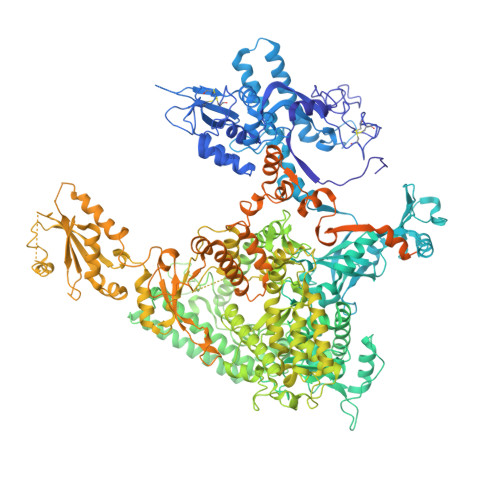
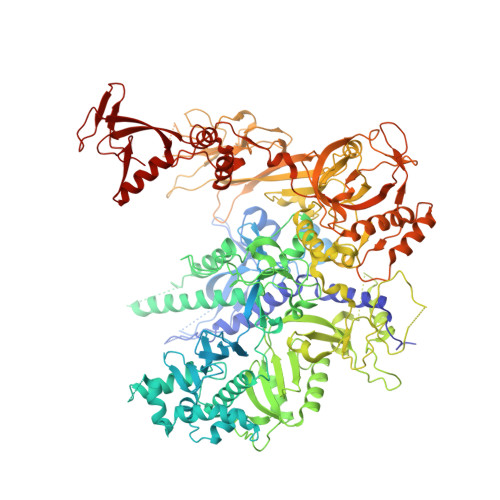
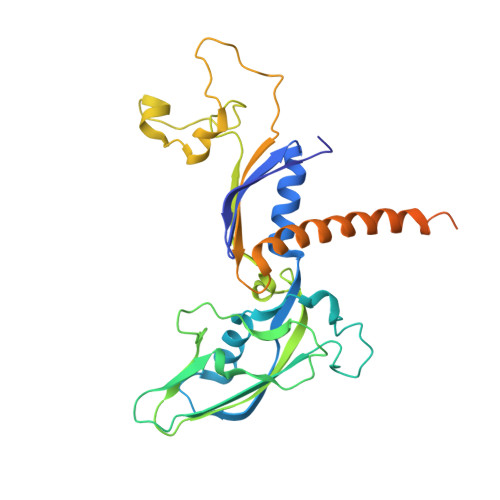
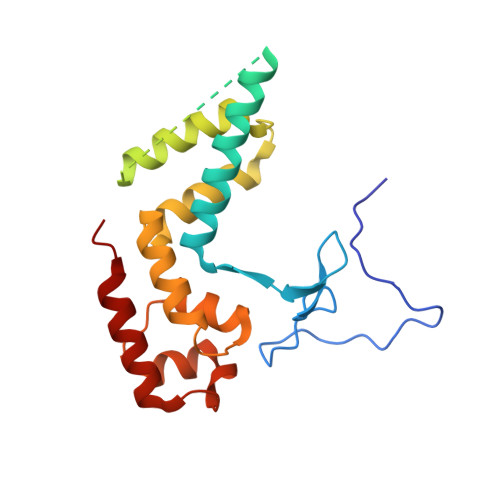
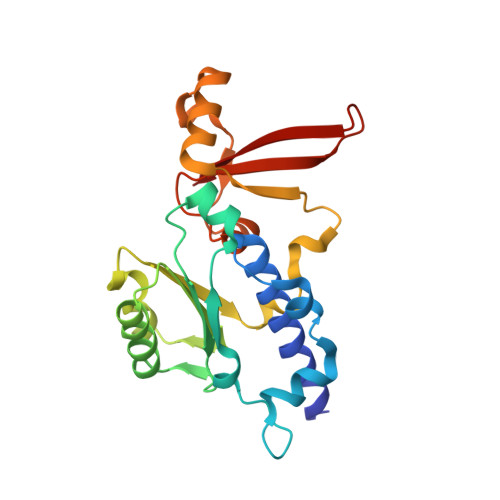
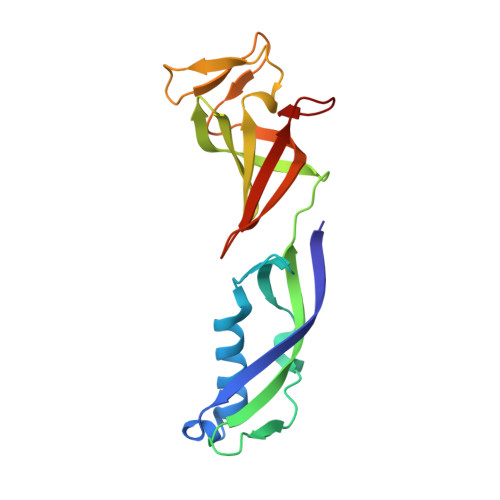
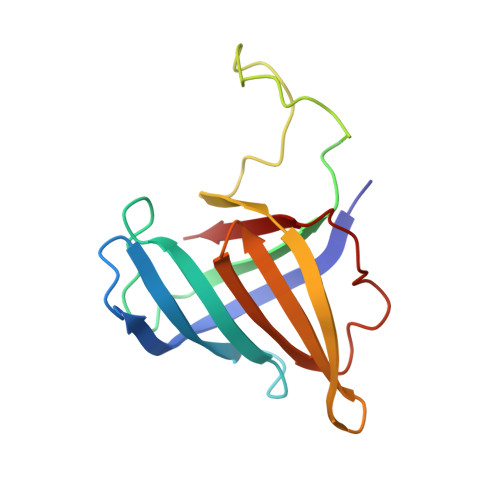
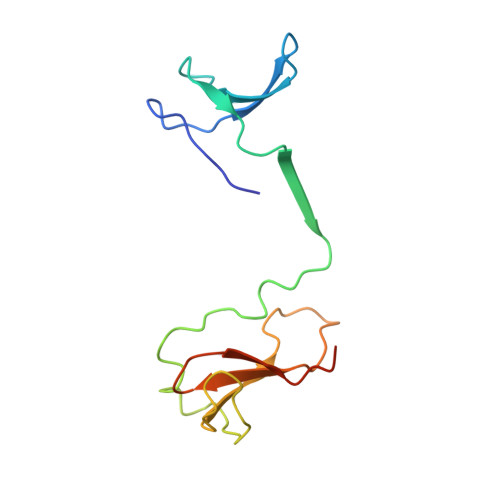
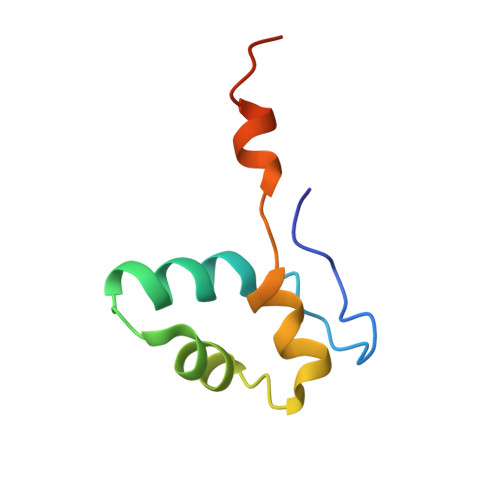
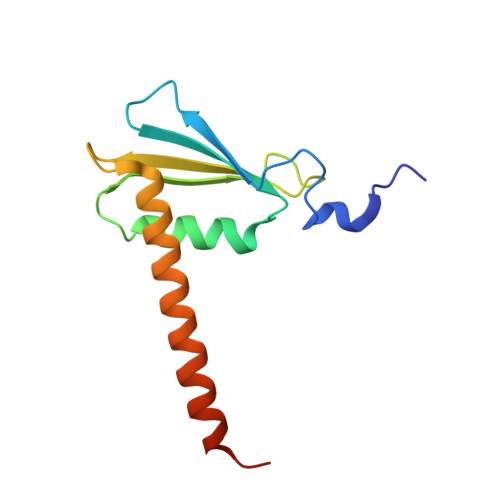
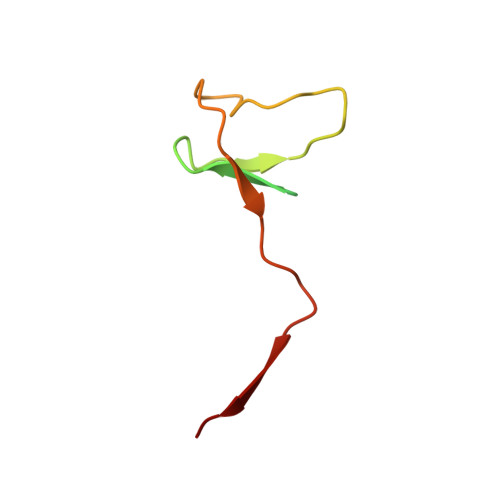
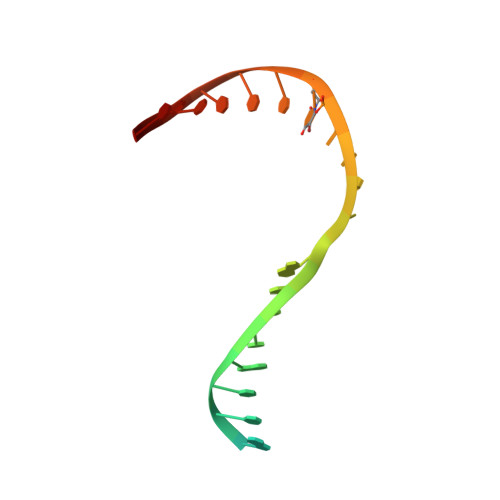
Mechanism of RNA polymerase II stalling by DNA alkylation.
Malvezzi, S., Farnung, L., Aloisi, C.M.N., Angelov, T., Cramer, P., Sturla, S.J.(2017) Proc Natl Acad Sci U S A 114: 12172-12177
- PubMed: 29087308
- DOI: https://doi.org/10.1073/pnas.1706592114
- Primary Citation of Related Structures:
5OT2 - PubMed Abstract:
Several anticancer agents that form DNA adducts in the minor groove interfere with DNA replication and transcription to induce apoptosis. Therapeutic resistance can occur, however, when cells are proficient in the removal of drug-induced damage. Acylfulvenes are a class of experimental anticancer agents with a unique repair profile suggesting their capacity to stall RNA polymerase (Pol) II and trigger transcription-coupled nucleotide excision repair. Here we show how different forms of DNA alkylation impair transcription by RNA Pol II in cells and with the isolated enzyme and unravel a mode of RNA Pol II stalling that is due to alkylation of DNA in the minor groove. We incorporated a model for acylfulvene adducts, the stable 3-deaza-3-methoxynaphtylethyl-adenosine analog (3d-Napht-A), and smaller 3-deaza-adenosine analogs, into DNA oligonucleotides to assess RNA Pol II transcription elongation in vitro. RNA Pol II was strongly blocked by a 3d-Napht-A analog but bypassed smaller analogs. Crystal structure analysis revealed that a DNA base containing 3d-Napht-A can occupy the +1 templating position and impair closing of the trigger loop in the Pol II active center and polymerase translocation into the next template position. These results show how RNA Pol II copes with minor-groove DNA alkylation and establishes a mechanism for drug resistance.
- Department of Health Sciences and Technology, ETH Zurich, 8092 Zurich, Switzerland.
Organizational Affiliation: