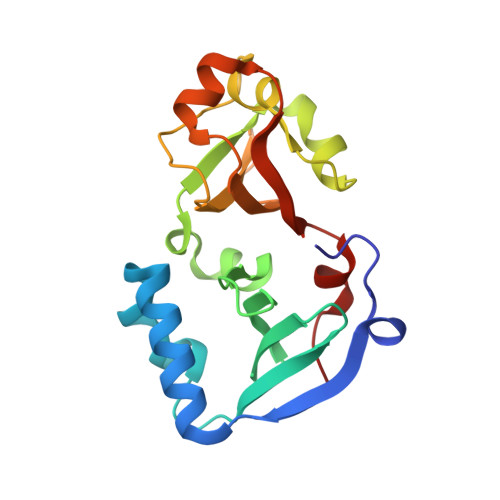
DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation.
Ahmed, Y.L., Schleich, S., Bohlen, J., Mandel, N., Simon, B., Sinning, I., Teleman, A.A.(2018) PLoS Biol 16: e2005160-e2005160
- PubMed: 29889857
- DOI: https://doi.org/10.1371/journal.pbio.2005160
- Primary Citation of Related Structures:
5ONS - PubMed Abstract:
The succession of molecular events leading to eukaryotic translation reinitiation-whereby ribosomes terminate translation of a short open reading frame (ORF), resume scanning, and then translate a second ORF on the same mRNA-is not well understood. Density-regulated reinitiation and release factor (DENR) and multiple copies in T-cell lymphoma-1 (MCTS1) are implicated in promoting translation reinitiation both in vitro in translation extracts and in vivo. We present here the crystal structure of MCTS1 bound to a fragment of DENR. Based on this structure, we identify and experimentally validate that DENR residues Glu42, Tyr43, and Tyr46 are important for MCTS1 binding and that MCTS1 residue Phe104 is important for tRNA binding. Mutation of these residues reveals that DENR-MCTS1 dimerization and tRNA binding are both necessary for DENR and MCTS1 to promote translation reinitiation in human cells. These findings thereby link individual residues of DENR and MCTS1 to specific molecular functions of the complex. Since DENR-MCTS1 can bind tRNA in the absence of the ribosome, this suggests the DENR-MCTS1 complex could recruit tRNA to the ribosome during reinitiation analogously to the eukaryotic initiation factor 2 (eIF2) complex in cap-dependent translation.
- Heidelberg University Biochemistry Center (BZH), Heidelberg, Germany.
Organizational Affiliation: