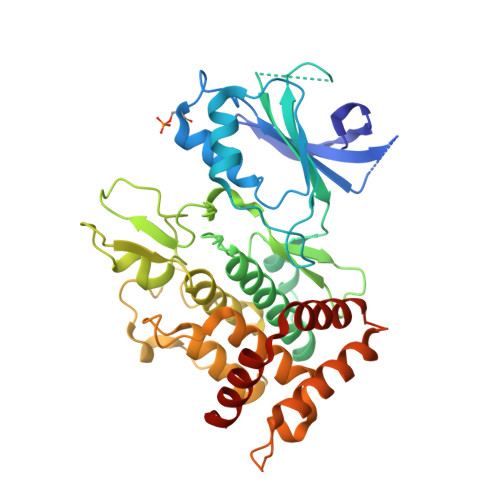
Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Kumar, A., Tamjar, J., Waddell, A.D., Woodroof, H.I., Raimi, O.G., Shaw, A.M., Peggie, M., Muqit, M.M., van Aalten, D.M.(2017) Elife 6
- PubMed: 28980524
- DOI: https://doi.org/10.7554/eLife.29985
- Primary Citation of Related Structures:
5OAT - PubMed Abstract:
Mutations in the human kinase PINK1 (hPINK1) are associated with autosomal recessive early-onset Parkinson's disease (PD). hPINK1 activates Parkin E3 ligase activity, involving phosphorylation of ubiquitin and the Parkin ubiquitin-like (Ubl) domain via as yet poorly understood mechanisms. hPINK1 is unusual amongst kinases due to the presence of three loop insertions of unknown function. We report the structure of Tribolium castaneum PINK1 ( Tc PINK1), revealing several unique extensions to the canonical protein kinase fold. The third insertion, together with autophosphorylation at residue Ser205, contributes to formation of a bowl-shaped binding site for ubiquitin. We also define a novel structural element within the second insertion that is held together by a distal loop that is critical for Tc PINK1 activity. The structure of Tc PINK1 explains how PD-linked mutations that lie within the kinase domain result in hPINK1 loss-of-function and provides a platform for the exploration of small molecule modulators of hPINK1.
- Division of Gene Regulation and Expression, University of Dundee, Dundee, United Kingdom.
Organizational Affiliation: