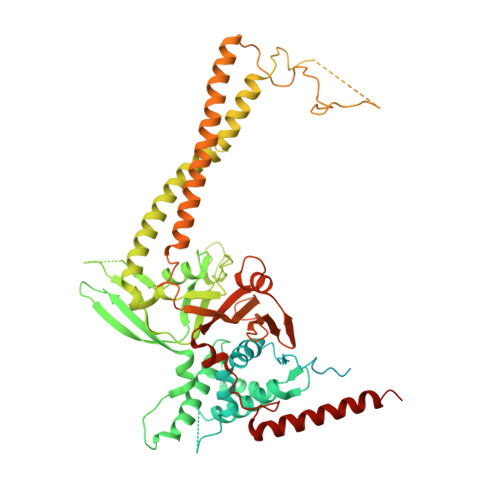
A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25.
Liu, B., Sureda-Gomez, M., Zhen, Y., Amador, V., Reverter, D.(2018) Nat Commun 9: 4973-4973
- PubMed: 30478318
- DOI: https://doi.org/10.1038/s41467-018-07510-5
- Primary Citation of Related Structures:
5O71 - PubMed Abstract:
USP25 deubiquitinating enzyme is a key member of the ubiquitin system, which acts as a positive regulator of the Wnt/β-catenin signaling by promoting the deubiquitination and stabilization of tankyrases. USP25 is characterized by the presence of a long insertion in the middle of the conserved catalytic domain. The crystal structure of USP25 displays an unexpected homotetrameric quaternary assembly that is directly involved in the inhibition of its enzymatic activity. The tetramer is assembled by the association of two dimers and includes contacts between the coiled-coil insertion domain and the ubiquitin-binding pocket at the catalytic domain, revealing a distinctive autoinhibitory mechanism. Biochemical and kinetic assays with dimer, tetramer and truncation constructs of USP25 support this mechanism, displaying higher catalytic activity in the dimer assembly. Moreover, the high stabilization of tankyrases in cultured cells by ectopic expression of a constitutive dimer of USP25 supports a biological relevance of this tetramerization/inhibition mechanism.
- Institut de Biotecnologia i de Biomedicina (IBB), Universitat Autònoma de Barcelona, Bellaterra, 08193, Spain.
Organizational Affiliation: