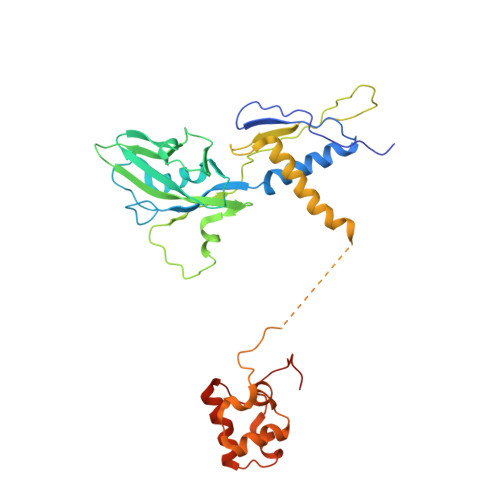
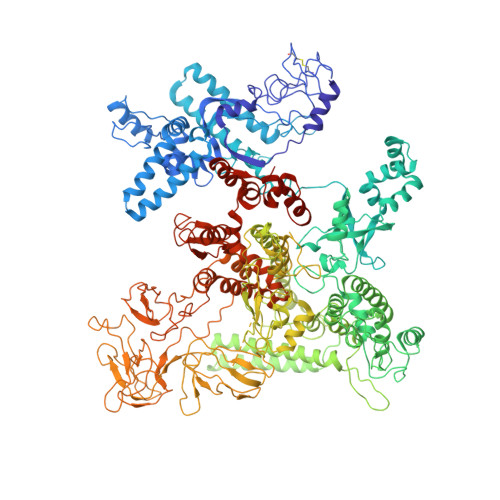
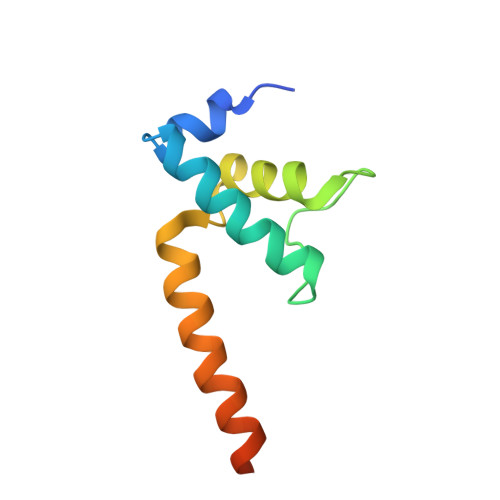
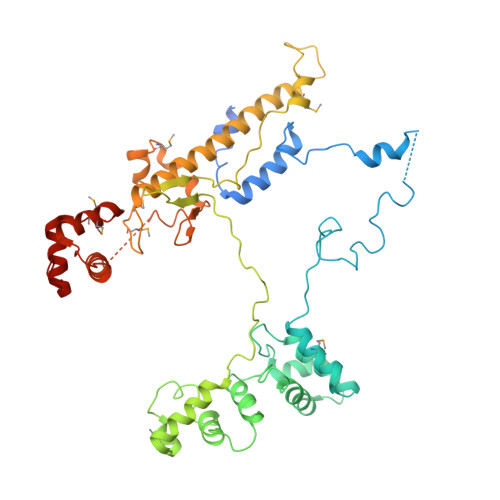
TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Yang, Y., Darbari, V.C., Zhang, N., Lu, D., Glyde, R., Wang, Y.P., Winkelman, J.T., Gourse, R.L., Murakami, K.S., Buck, M., Zhang, X.(2015) Science 349: 882-885
- PubMed: 26293966
- DOI: https://doi.org/10.1126/science.aab1478
- Primary Citation of Related Structures:
5EZK, 5NWT - PubMed Abstract:
Transcription by RNA polymerase (RNAP) in bacteria requires specific promoter recognition by σ factors. The major variant σ factor (σ(54)) initially forms a transcriptionally silent complex requiring specialized adenosine triphosphate-dependent activators for initiation. Our crystal structure of the 450-kilodalton RNAP-σ(54) holoenzyme at 3.8 angstroms reveals molecular details of σ(54) and its interactions with RNAP. The structure explains how σ(54) targets different regions in RNAP to exert its inhibitory function. Although σ(54) and the major σ factor, σ(70), have similar functional domains and contact similar regions of RNAP, unanticipated differences are observed in their domain arrangement and interactions with RNAP, explaining their distinct properties. Furthermore, we observe evolutionarily conserved regulatory hotspots in RNAPs that can be targeted by a diverse range of mechanisms to fine tune transcription.
- Centre for Structural Biology, Imperial College London, South Kensington SW7 2AZ, UK. State Key Laboratory of Protein and Plant Gene Research, College of Life Sciences, Peking University, China.
Organizational Affiliation: