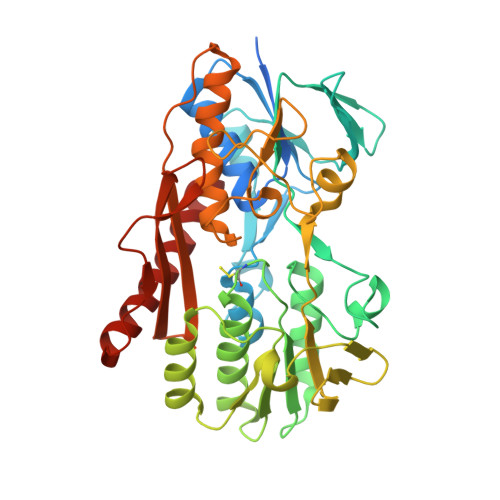
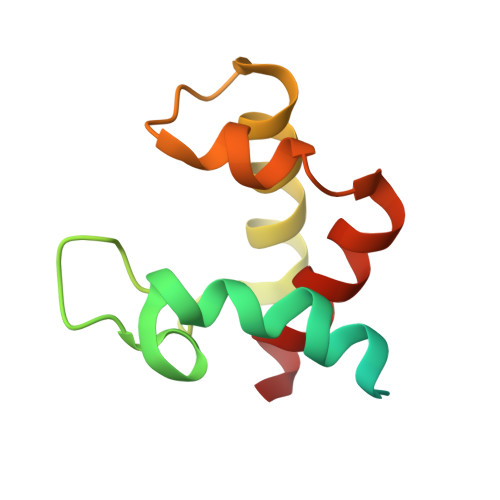
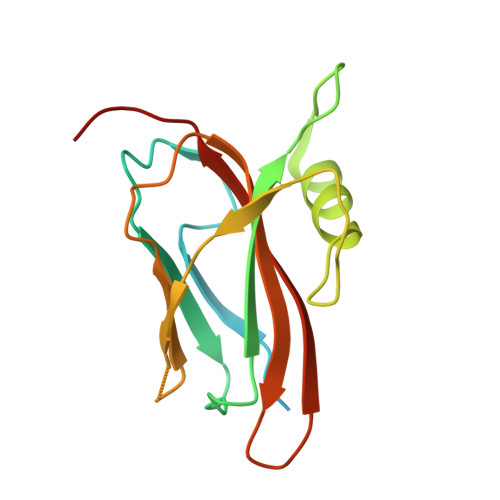
Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Osipov, E.M., Lilina, A.V., Tsallagov, S.I., Safonova, T.N., Sorokin, D.Y., Tikhonova, T.V., Popov, V.O.(2018) Acta Crystallogr D Struct Biol 74: 632-642
- PubMed: 29968673
- DOI: https://doi.org/10.1107/S2059798318005648
- Primary Citation of Related Structures:
5N1T - PubMed Abstract:
Flavocytochrome c sulfide dehydrogenase from Thioalkalivibrio paradoxus (TpFCC) is a heterodimeric protein consisting of flavin- and monohaem c-binding subunits. TpFCC was co-purified and co-crystallized with the dimeric copper-binding protein TpCopC. The structure of the TpFCC-(TpCopC) 2 complex was determined by X-ray diffraction at 2.6 Å resolution. The flavin-binding subunit of TpFCC is structurally similar to those determined previously, and the structure of the haem-binding subunit is similar to that of the N-terminal domain of dihaem FCCs. According to classification based on amino-acid sequence, TpCopC belongs to a high-affinity CopC subfamily characterized by the presence of a conserved His1-Xxx-His3 motif at the N-terminus. Apparently, a unique α-helix which is present in each monomer of TpCopC at the interface with TpFCC plays a key role in complex formation. The structure of the copper-binding site in TpCopC is similar to those in other known CopC structures. His3 is not involved in binding to the copper ion and is 6-7 Å away from this ion. Therefore, the His1-Xxx-His3 motif cannot be considered to be a key factor in the high affinity of CopC for copper(II) ions. It is suggested that the TpFCC-(TpCopC) 2 heterotetramer may be a component of a large periplasmic complex that is responsible for thiocyanate metabolism.
- Research Center of Biotechnology of the Russian Academy of Sciences, 33 Leninsky Avenue, Building 2, Moscow 119071, Russian Federation.
Organizational Affiliation: