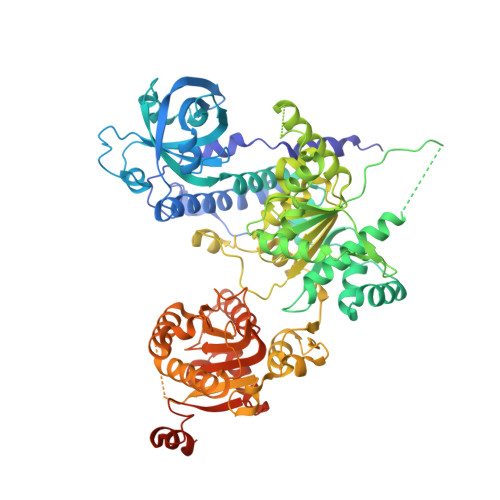
Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family.
Leonaite, B., Han, Z., Basquin, J., Bonneau, F., Libri, D., Porrua, O., Conti, E.(2017) EMBO J 36: 1590-1604
- PubMed: 28408439
- DOI: https://doi.org/10.15252/embj.201696174
- Primary Citation of Related Structures:
5MZN - PubMed Abstract:
The superfamily 1B (SF1B) helicase Sen1 is an essential protein that plays a key role in the termination of non-coding transcription in yeast. Here, we identified the ~90 kDa helicase core of Saccharomyces cerevisiae Sen1 as sufficient for transcription termination in vitro and determined the corresponding structure at 1.8 Å resolution. In addition to the catalytic and auxiliary subdomains characteristic of the SF1B family, Sen1 has a distinct and evolutionarily conserved structural feature that "braces" the helicase core. Comparative structural analyses indicate that the "brace" is essential in shaping a favorable conformation for RNA binding and unwinding. We also show that subdomain 1C (the "prong") is an essential element for 5'-3' unwinding and for Sen1-mediated transcription termination in vitro Finally, yeast Sen1 mutant proteins mimicking the disease forms of the human orthologue, senataxin, show lower capacity of RNA unwinding and impairment of transcription termination in vitro The combined biochemical and structural data thus provide a molecular model for the specificity of Sen1 in transcription termination and more generally for the unwinding mechanism of 5'-3' helicases.
- Max Planck Institute of Biochemistry, Munich, Germany.
Organizational Affiliation: