Efficient conversion of alkenes to chlorohydrins by a Ru-based artificial enzyme
Lopez, S., Rondot, L., Cavazza, C., Iannello, M., Boeri-Erba, E., Burzlaff, N., Strinitz, N., Jorge-Robin, A., Marchi-Delapierre, C., Menage, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nickel-binding periplasmic protein | 502 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: nikA, b3476, JW3441 | 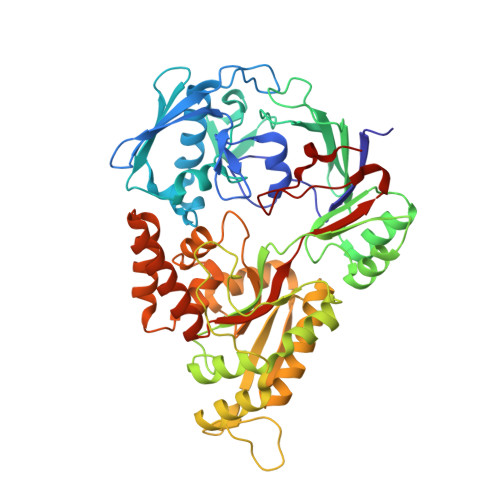 | |
UniProt | |||||
Find proteins for P33590 (Escherichia coli (strain K12)) Explore P33590 Go to UniProtKB: P33590 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P33590 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDT Query on EDT | K [auth A] | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID C10 H16 N2 O8 KCXVZYZYPLLWCC-UHFFFAOYSA-N |  | ||
| 6RP Query on 6RP | AA [auth B] | bis(pyrzol-1-yl)acetate scorpionate C8 H8 N4 O2 NJDSSVBTTVUKHM-UHFFFAOYSA-N |  | ||
| RU Query on RU | Y [auth B] | RUTHENIUM ION Ru BPEVHDGLPIIAGH-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | F [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | DA [auth B] G [auth A] GA [auth B] H [auth A] I [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | C [auth A] D [auth A] E [auth A] FA [auth B] L [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| FE Query on FE | J [auth A] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| CL Query on CL | EA [auth B], Z [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| CMO Query on CMO | BA [auth B], CA [auth B] | CARBON MONOXIDE C O UGFAIRIUMAVXCW-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86.303 | α = 90 |
| b = 93.606 | β = 90 |
| c = 124.211 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| French National Research Agency | France | ANR-14-CE06-0005-01 |