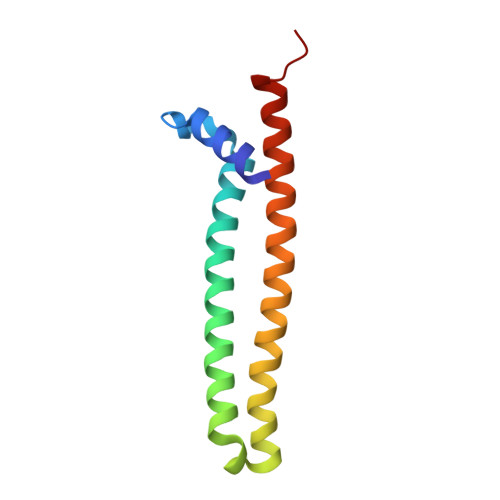
The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
Ormaza, G., Rodriguez, J.A., Ibanez de Opakua, A., Merino, N., Villate, M., Gorrono, I., Rabano, M., Palmero, I., Vilaseca, M., Kypta, R., Vivanco, M.D.M., Rojas, A.L., Blanco, F.J.(2019) J Mol Biology 431: 2298-2319
- PubMed: 31026448
- DOI: https://doi.org/10.1016/j.jmb.2019.04.018
- Primary Citation of Related Structures:
5ME8, 5MTO - PubMed Abstract:
The INhibitor of Growth (ING) family of tumor suppressors regulates the transcriptional state of chromatin by recruiting remodeling complexes to sites with histone H3 trimethylated at lysine 4 (H3K4me3). This modification is recognized by the plant homeodomain (PHD) present at the C-terminus of the five ING proteins. ING5 facilitates histone H3 acetylation by the HBO1 complex, and also H4 acetylation by the MOZ/MORF complex. We show that ING5 forms homodimers through its N-terminal domain, which folds independently into an elongated coiled-coil structure. The central region of ING5, which contains the nuclear localization sequence, is flexible and disordered, but it binds dsDNA with micromolar affinity. NMR analysis of the full-length protein reveals that the two PHD fingers of the dimer are chemically equivalent and independent of the rest of the molecule, and they bind H3K4me3 in the same way as the isolated PHD. We have observed that ING5 can form heterodimers with the highly homologous ING4, and that two of three primary tumor-associated mutants in the N-terminal domain strongly destabilize the coiled-coil structure. They also affect cell proliferation and cell cycle phase distribution, suggesting a driver role in cancer progression.
- CIC bioGUNE, Parque Tecnológico de Bizkaia, 48160 Derio, Spain.
Organizational Affiliation: