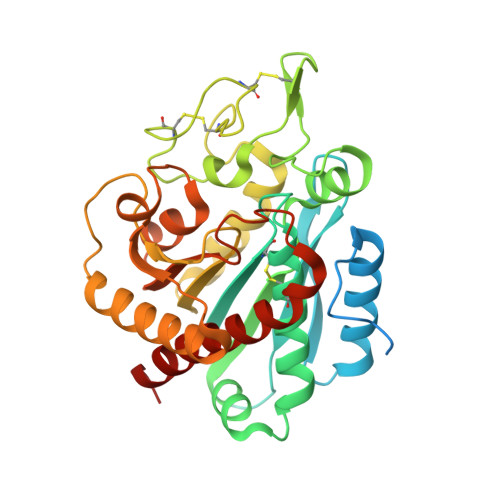
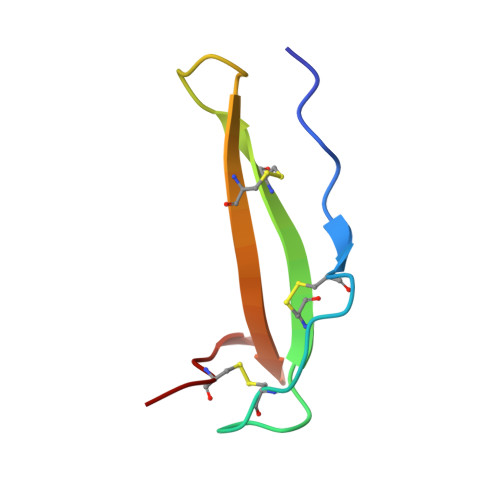
Crystal structure and mechanism of human carboxypeptidase O: Insights into its specific activity for acidic residues.
Garcia-Guerrero, M.C., Garcia-Pardo, J., Berenguer, E., Fernandez-Alvarez, R., Barfi, G.B., Lyons, P.J., Aviles, F.X., Huber, R., Lorenzo, J., Reverter, D.(2018) Proc Natl Acad Sci U S A 115: E3932-E3939
- PubMed: 29636417
- DOI: https://doi.org/10.1073/pnas.1803685115
- Primary Citation of Related Structures:
5MRV - PubMed Abstract:
Human metallocarboxypeptidase O (hCPO) is a recently discovered digestive enzyme localized to the apical membrane of intestinal epithelial cells. Unlike pancreatic metallocarboxypeptidases, hCPO is glycosylated and produced as an active enzyme with distinctive substrate specificity toward C-terminal (C-t) acidic residues. Here we present the crystal structure of hCPO at 1.85-Å resolution, both alone and in complex with a carboxypeptidase inhibitor (NvCI) from the marine snail Nerita versicolor The structure provides detailed information regarding determinants of enzyme specificity, in particular Arg275, placed at the bottom of the substrate-binding pocket. This residue, located at "canonical" position 255, where it is Ile in human pancreatic carboxypeptidases A1 (hCPA1) and A2 (hCPA2) and Asp in B (hCPB), plays a dominant role in determining the preference of hCPO for acidic C-t residues. Site-directed mutagenesis to Asp and Ala changes the specificity to C-t basic and hydrophobic residues, respectively. The single-site mutants thus faithfully mimic the enzymatic properties of CPB and CPA, respectively. hCPO also shows a preference for Glu over Asp, probably as a consequence of a tighter fitting of the Glu side chain in its S1' substrate-binding pocket. This unique preference of hCPO, together with hCPA1, hCPA2, and hCPB, completes the array of C-t cleavages enabling the digestion of the dietary proteins within the intestine. Finally, in addition to activity toward small synthetic substrates and peptides, hCPO can also trim C-t extensions of proteins, such as epidermal growth factor, suggesting a role in the maturation and degradation of growth factors and bioactive peptides.
- Institute for Biotechnology and Biomedicine, Universitat Autonoma de Barcelona, 08193 Bellaterra, Barcelona, Spain.
Organizational Affiliation: