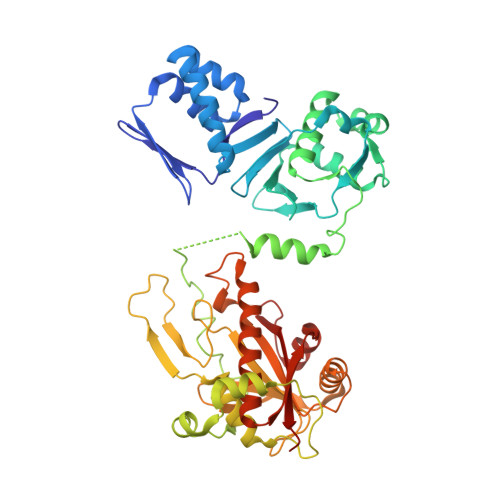
Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Bent, A.F., Mann, G., Houssen, W.E., Mykhaylyk, V., Duman, R., Thomas, L., Jaspars, M., Wagner, A., Naismith, J.H.(2016) Acta Crystallogr D Struct Biol 72: 1174-1180
- PubMed: 27841750
- DOI: https://doi.org/10.1107/S2059798316015850
- Primary Citation of Related Structures:
5LQ4 - PubMed Abstract:
Determination of protein crystal structures requires that the phases are derived independently of the observed measurement of diffraction intensities. Many techniques have been developed to obtain phases, including heavy-atom substitution, molecular replacement and substitution during protein expression of the amino acid methionine with selenomethionine. Although the use of selenium-containing methionine has transformed the experimental determination of phases it is not always possible, either because the variant protein cannot be produced or does not crystallize. Phasing of structures by measuring the anomalous diffraction from S atoms could in theory be almost universal since almost all proteins contain methionine or cysteine. Indeed, many structures have been solved by the so-called native sulfur single-wavelength anomalous diffraction (S-SAD) phasing method. However, the anomalous effect is weak at the wavelengths where data are normally recorded (between 1 and 2 Å) and this limits the potential of this method to well diffracting crystals. Longer wavelengths increase the strength of the anomalous signal but at the cost of increasing air absorption and scatter, which degrade the precision of the anomalous measurement, consequently hindering phase determination. A new instrument, the long-wavelength beamline I23 at Diamond Light Source, was designed to work at significantly longer wavelengths compared with standard synchrotron beamlines in order to open up the native S-SAD method to projects of increasing complexity. Here, the first novel structure, that of the oxidase domain involved in the production of the natural product patellamide, solved on this beamline is reported using data collected to a resolution of 3.15 Å at a wavelength of 3.1 Å. The oxidase is an example of a protein that does not crystallize as the selenium variant and for which no suitable homology model for molecular replacement was available. Initial attempts collecting anomalous diffraction data for native sulfur phasing on a standard macromolecular crystallography beamline using a wavelength of 1.77 Å did not yield a structure. The new beamline thus has the potential to facilitate structure determination by native S-SAD phasing for what would previously have been regarded as very challenging cases with modestly diffracting crystals and low sulfur content.
- BSRC, University of St Andrews, North Haugh, St Andrews, Fife KY16 9ST, Scotland.
Organizational Affiliation: