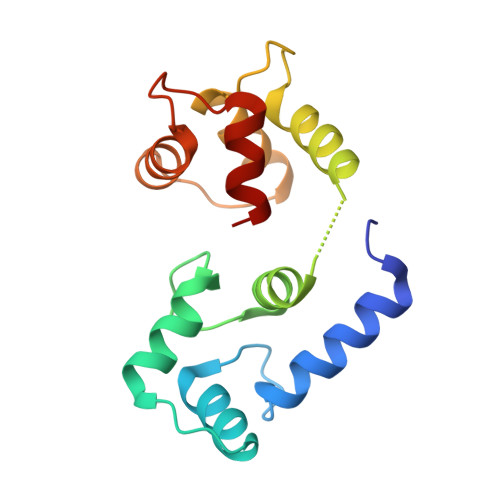
Structure of the STRA6 receptor for retinol uptake.
Chen, Y., Clarke, O.B., Kim, J., Stowe, S., Kim, Y.K., Assur, Z., Cavalier, M., Godoy-Ruiz, R., von Alpen, D.C., Manzini, C., Blaner, W.S., Frank, J., Quadro, L., Weber, D.J., Shapiro, L., Hendrickson, W.A., Mancia, F.(2016) Science 353
- PubMed: 27563101
- DOI: https://doi.org/10.1126/science.aad8266
- Primary Citation of Related Structures:
5K8Q, 5SY1 - PubMed Abstract:
Vitamin A homeostasis is critical to normal cellular function. Retinol-binding protein (RBP) is the sole specific carrier in the bloodstream for hydrophobic retinol, the main form in which vitamin A is transported. The integral membrane receptor STRA6 mediates cellular uptake of vitamin A by recognizing RBP-retinol to trigger release and internalization of retinol. We present the structure of zebrafish STRA6 determined to 3.9-angstrom resolution by single-particle cryo-electron microscopy. STRA6 has one intramembrane and nine transmembrane helices in an intricate dimeric assembly. Unexpectedly, calmodulin is bound tightly to STRA6 in a noncanonical arrangement. Residues involved with RBP binding map to an archlike structure that covers a deep lipophilic cleft. This cleft is open to the membrane, suggesting a possible mode for internalization of retinol through direct diffusion into the lipid bilayer.
- Department of Physiology and Cellular Biophysics, Columbia University, New York, NY 10032, USA.
Organizational Affiliation: